All
Company news
Industry insights
Press releases
Product updates


Tailored workflows in discovery research: Challenges, benefits, and how to implement them today

Reducing the barriers for immune repertoire research: An in-depth interview with Dr. Felix Breden

ENPICOM secures financing from BOP Capital, BOM Brabant Ventures and brings Dr. Colby Souders on board as a new investor

Simulating adaptive immune receptor repertoire sequencing runs with InSilicoSeq

ENPICOM partners with Erasmus University Medical Center in groundbreaking effort to discover nanobodies against cancer

ENPICOM and Carterra advance antibody screening by integrating biophysical characterization and repertoire sequencing analysis
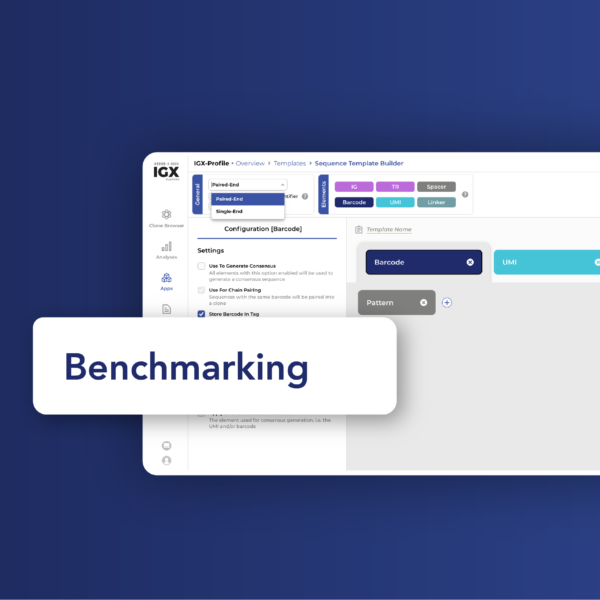
In pursuit of precision: Validating IGX-Profile against a benchmark Rep-Seq dataset

Navigating the immune repertoire: A deep dive into applications, workflows, and new sequencing chemistries

From sample to discovery: Applications, challenges, and the path ahead in full-service repertoire sequencing

Adopting a user-centered approach to biotech software design and development
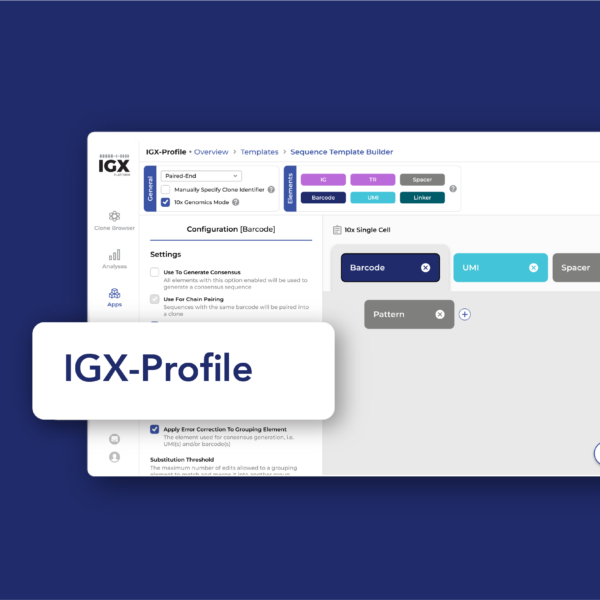
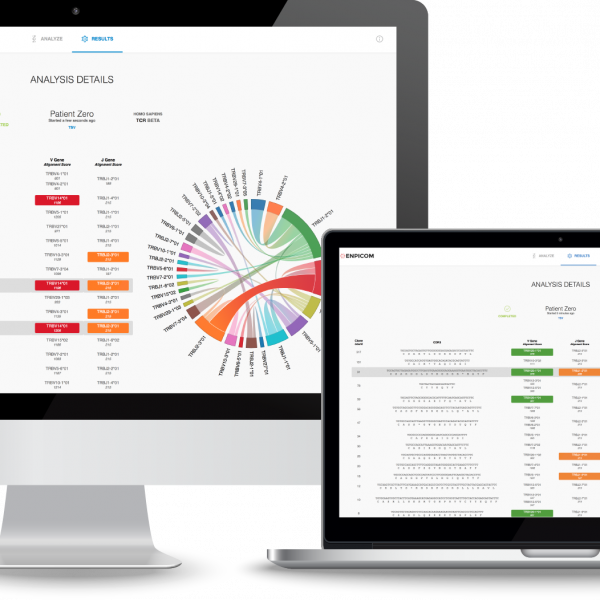
Accurate clone annotation for antibody and TCR discovery

ENPICOM secures extended financing from BOP Capital, BOM Brabant Ventures, Nextgen Ventures, and Arches Capital

ENPICOM announces CEO transition

Authentication in the IGX Platform

2022 in review: ENPICOM’s highlights

Efficiently using Amazon Web Services

Strength in numbers: Using next-generation sequencing with display technologies for antibody discovery

Vaccine development innovation & collaboration in action: The Inno4Vac project

The animal side of antibody discovery

ENPICOM receives ISO 27001:2013 certification
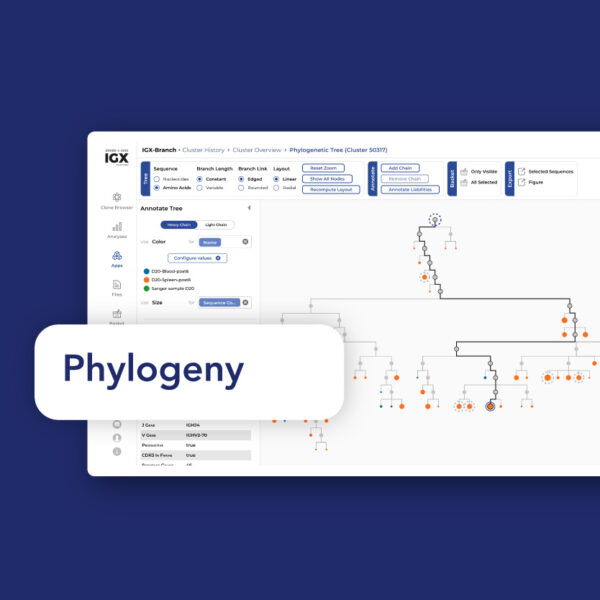
Phylogenetic analysis of BCR repertoires: Benefits, challenges, and considerations

Accurate developability assessments of antibody candidates with the IGX Platform

From a great idea to best in class SaaS Platform to improve immunotherapy discovery and development

Experiencing being an intern at ENPICOM

Paul van der Velde joins ENPICOM as Chief Operating Officer and Acting Chief Technology Officer

Meet IGX-Track: Our unique new App for seamless analysis of antibody display data

ENPICOM launches versatile display data analysis solution to accelerate antibody selection while maximizing precision

Genovac further enhances its antibody discovery services with the ENPICOM IGX Platform

ENPICOM increases speed and scalability in antibody discovery with Amazon Web Services (AWS)

ENPICOM secures Series B funding to scale up operations to meet the high demand for its antibody discovery SaaS Platform

2021 highlights: ENPICOM’s year in review

Meet IGX Platform 3.1: More flexibility and control for your seamless Rep-Seq analysis workflow

ENPICOM participates in Inno4vac, a European new public-private partnership to innovate vaccine development

From sample to discovery: Advancing your research with full-service immune repertoire sequencing & analysis

ENPICOM introduces a powerful liability prediction solution to de-risk antibody development

ENPICOM is featured among the top 50 fastest growing Data Science SMEs in the Netherlands

ENPICOM to integrate renowned structural antibody prediction tools developed at the University of Oxford

Empowering research teams: The path to accessible immune repertoire data

Zai Lab selects ENPICOM’s IGX Platform to leverage new NGS-powered antibody discovery workflows

The impact of high-throughput immune repertoire sequencing on the analysis of antibody responses: A conversation with Prof. Gunilla Karlsson Hedestam of Karolinska Institute

Webinar: Understanding the role of B cells in cancer through antibody repertoire analysis

Breaking the UX barrier: Towards user-focused biomedical software

ENPICOM and Viroclinics-DDL receive joint MIT R&D ZH subsidy to accelerate and improve antibody and vaccine discovery and development

ENPICOM joins the MIRIADE consortium

ENPICOM introduces an end-to-end solution for fast and efficient antibody discovery

ENPICOM and MiLaboratories join forces to power better Repertoire Sequencing data management and analysis workflows

ENPICOM services Neogene Therapeutics with its T cell repertoire sequencing data handling and analysis solution

ENPICOM is now a corporate sponsor of The Antibody Society
Single-cell sequencing: Industry trends and developments

University Health Network, Canada’s largest research hospital, selects ENPICOM’s IGX Platform to analyze and manage immune sequencing data

Gaining a deeper understanding of the immune system: A conversation with Prof. Dirk Busch and Dr. Kilian Schober of the Technical University of Munich

Fighting COVID-19 and transforming infectious disease research with immune repertoire sequencing
More data from a single cell: Advancing your immunology research with Repertoire Sequencing

ENPICOM receives a €1.13 million follow-on investment

ENPICOM deepens insights into the immune system enabling faster precision medicine development and pandemic prevention

ENPICOM and Cytura Therapeutics partner on early cancer detection

ENPICOM has been featured as one of the 20 cutting-edge tech companies of 2019

DDL diagnostic laboratory and ENPICOM join forces

ENPICOM launches unique immune sequencing data analysis platform

ENPICOM secures funding round to finalize IGX Platform development

Survey results: European immunotherapy companies have great concerns regarding increasing costs and long development timelines

ENPICOM sets a new standard for B cell receptor repertoire analysis

Towards a predictable combination therapy for multiple cancer types

ENPICOM team wins Microsoft Genomics Hackathon

Scientific poster from UKE/ENPICOM receives award at annual meeting of DGHO