ENPICOM Platform
Built to streamline collaboration between lab and data science teams, we provide a scalable, unified ecosystem to simplify data integration, AI model use, management, and training.
Empower lab scientists to easily access and apply AI models for analysis and decision making, driving innovation and accelerating breakthroughs.
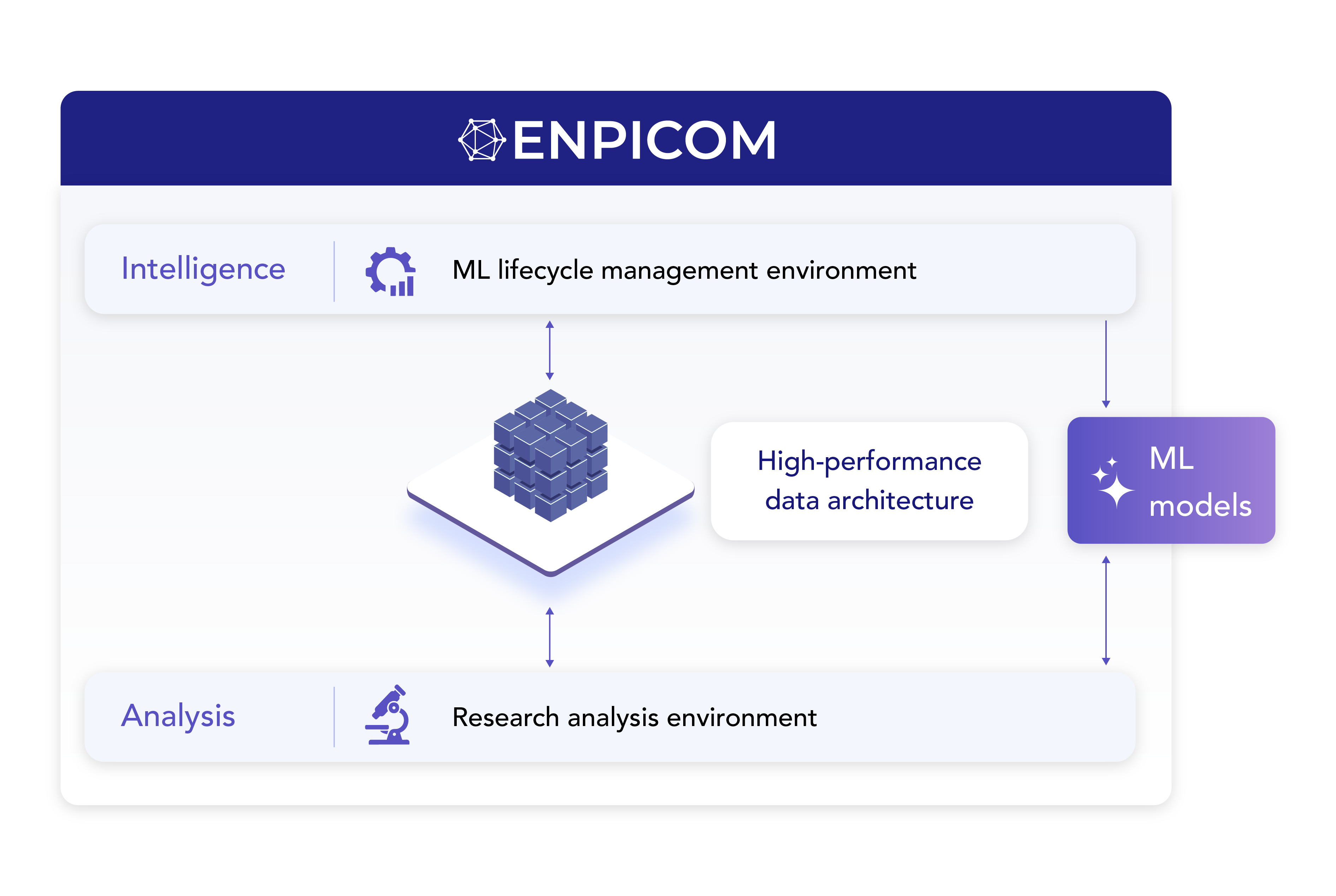
Unifying biologics research and AI effectively
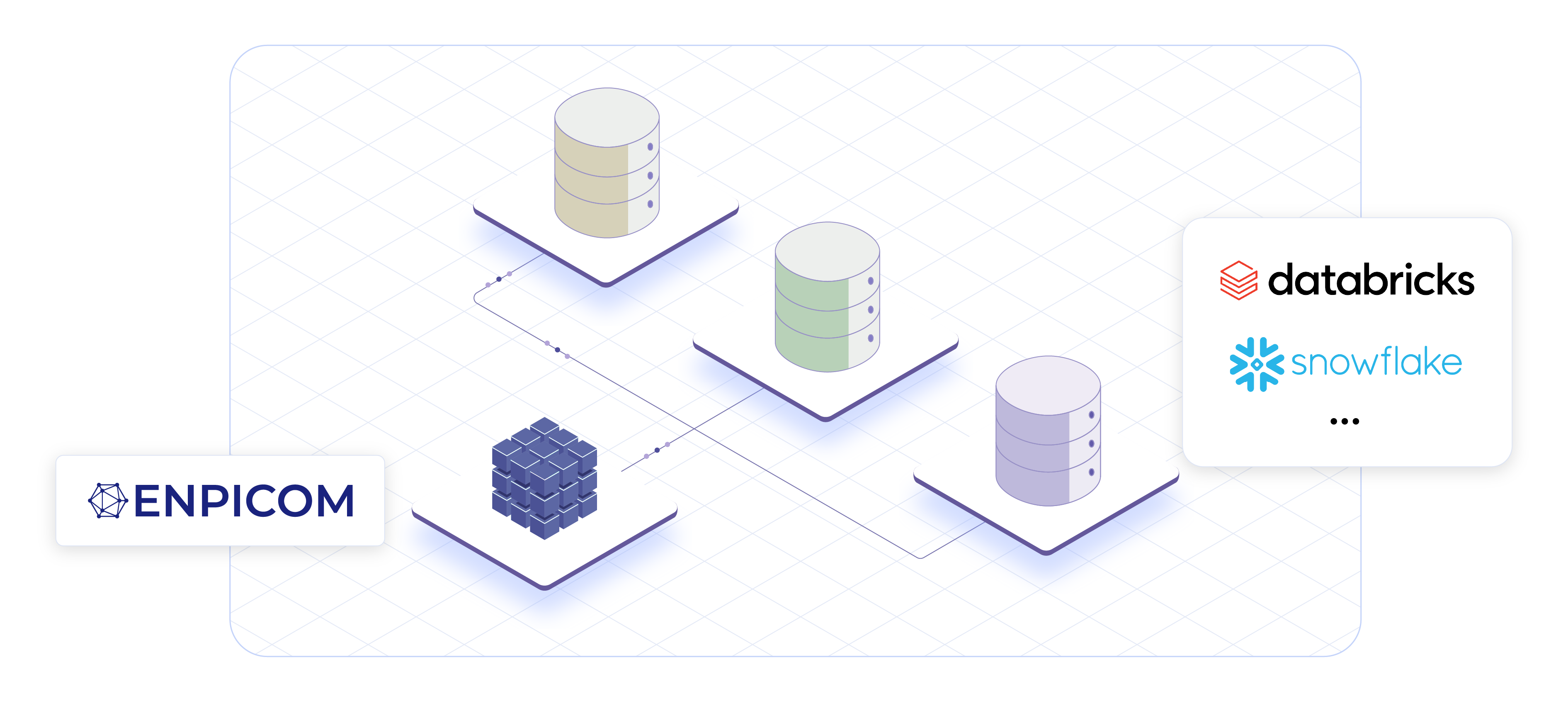
Scalable data foundation built for biologics
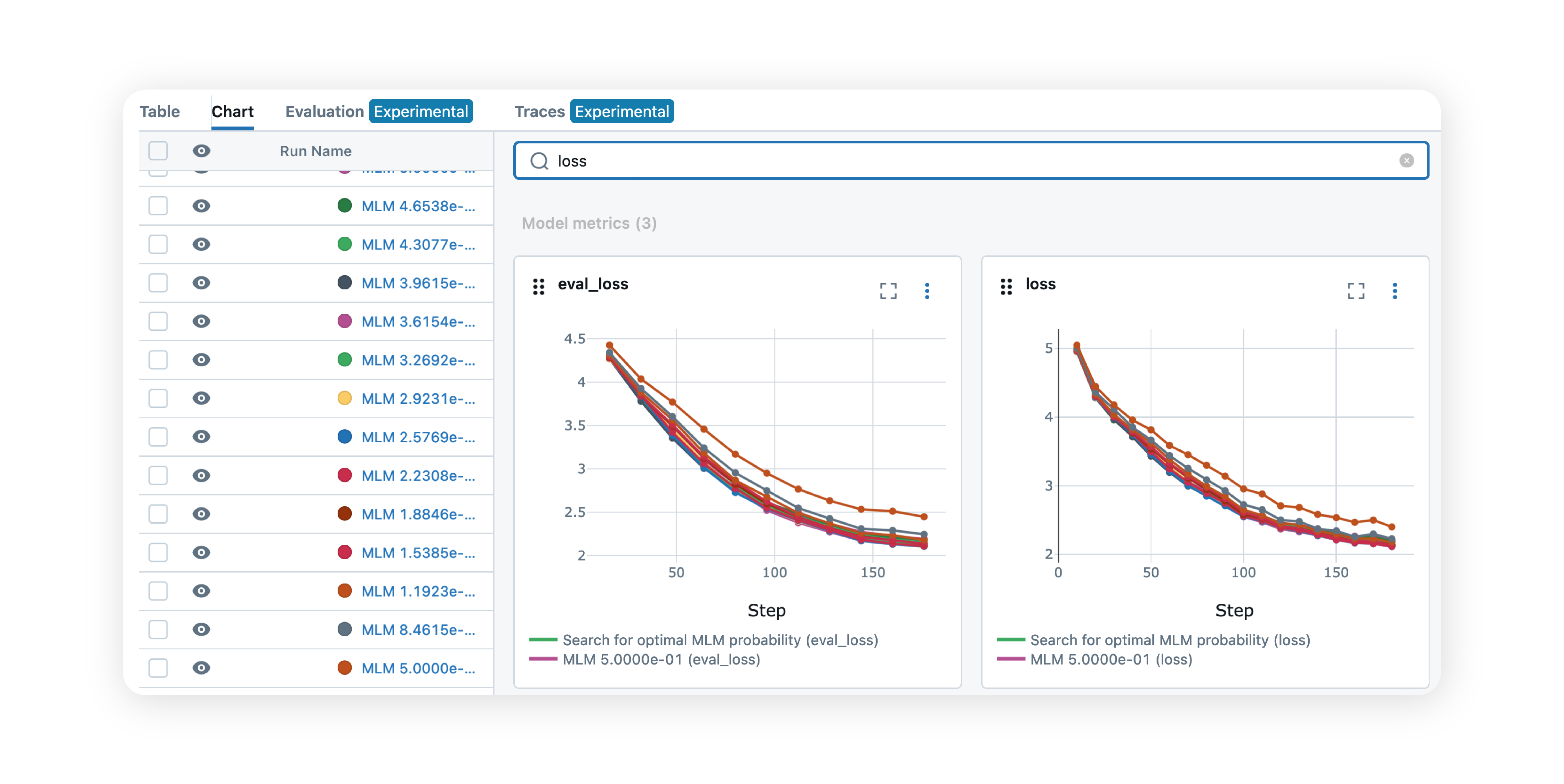
Effective AI-driven discovery depends on structured, high-throughput data. ENPICOM’s data architecture centralizes and harmonizes research data, seamlessly integrating it into your existing ecosystem. With our high-performance database, you can run complex queries on hundreds of millions of sequences in seconds, not days – helping to eliminate data silos and unlocking the full potential of your data.
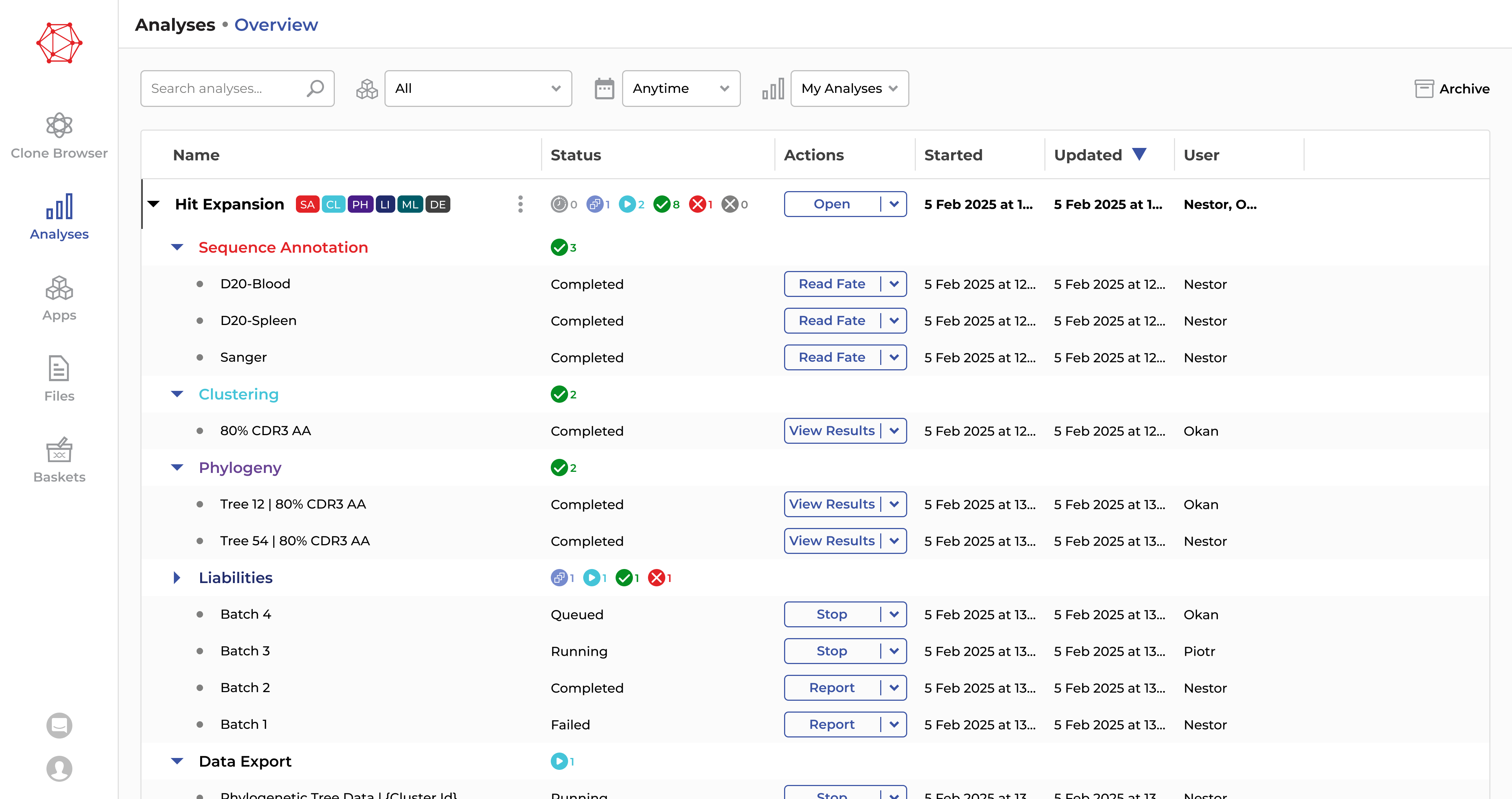
Automated analysis workflows
Streamline biologics discovery with automated, no-code analysis workflows. Our solution enables scientists to intuitively navigate complex datasets, driving faster and more accurate discoveries across all major discovery and engineering workflows.
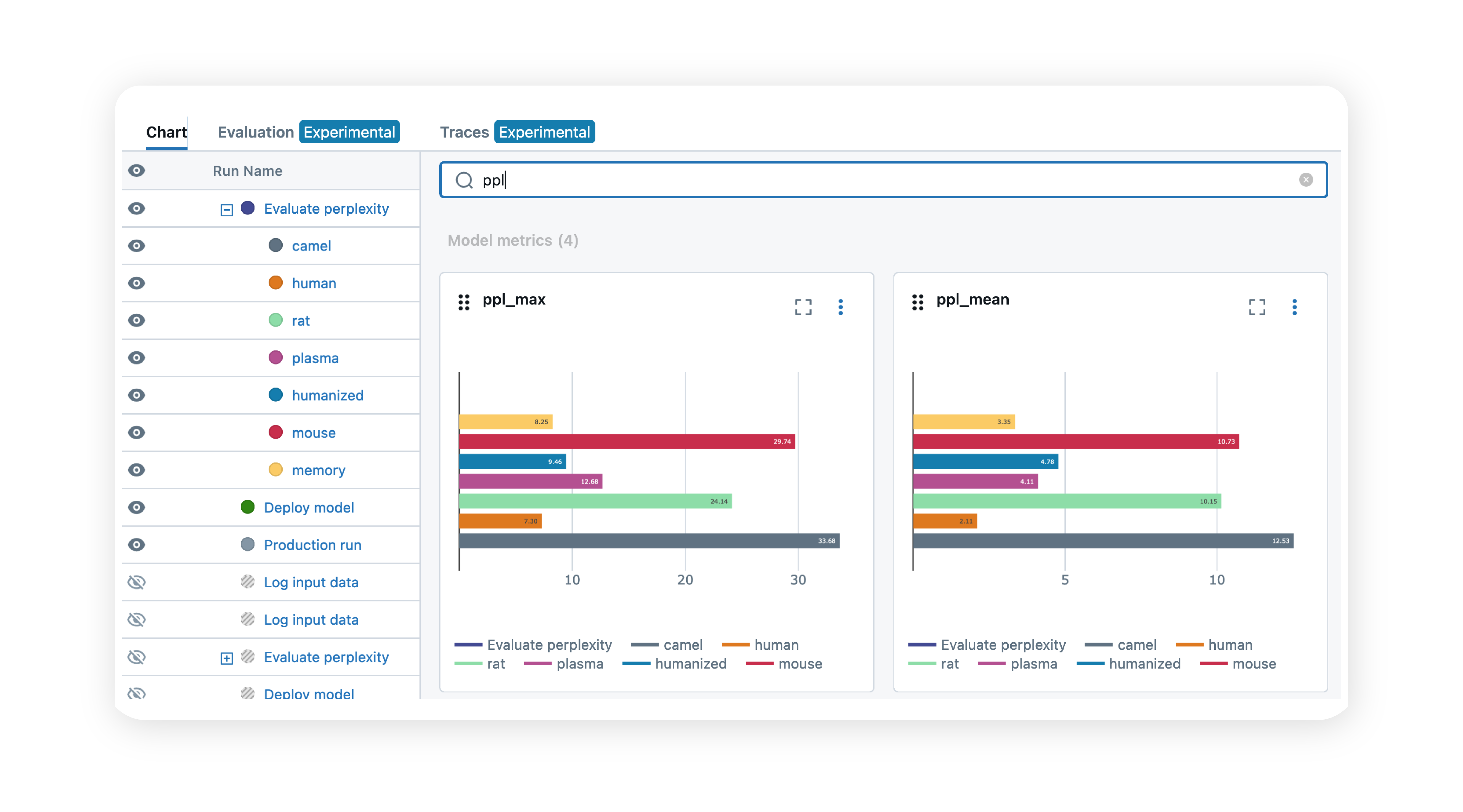
Streamline MLOps and easily deploy models
By streamlining ML model management, training, and deployment, we provide a robust infrastructure for MLOps and model adoption. This enables data scientists to focus on developing and perfecting models, ensuring they can be quickly deployed and tested and validated in real-world campaigns.
Truly unified experience
Empower your teams to collaborate effortlessly, streamline workflows, integrate AI, and accelerate biologics research — all within a single, fully integrated ecosystem that works in harmony to support every stage of your discovery and engineering journey.
Tailored discovery, delivered fast
Off-the-shelf solutions may not always meet the specific requirements of your discovery platforms and protocols. The journey to the discovery of successful therapeutics is as unique as your discovery platforms. That’s why we offer a fast and personalized approach to tailor analysis workflows to your needs. Say goodbye to one-size-fits-all approaches and enjoy a bespoke experience that caters to your specific requirements.
With deep expertise in discovery in-vitro and in-silico platforms, sequencing technologies, and data science, our team can rapidly tailor solutions to fit your unique needs and discovery environment. We ensure seamless integration of custom workflows into our scalable platform, providing continuous support and adapting to your evolving project requirements for lasting success.
Rapid customization
Work hand-in-hand with our experts to swiftly tailor solutions to your exact needs, keeping your project on track and ahead of the competition.
Standardization
Define and implement processes and SOPs to ensure efficacy, consistency, and speed across your organization.
Automation and simplicity
Seamlessly automate your workflows, eliminating repetitive tasks and streamlining your entire discovery process.
Effortless integration
Integrate the ENPICOM Platform into your discovery environment using APIs to align with your discovery goals and boost efficiency.
Unparalleled expertise
Benefit from our extensive experience in therapeutics discovery and bioinformatics, as our highly skilled engineers tackle even the most complex customization challenges.
Continuous support

Streamline all your teams and tools in biologics research
- Support for large-scale AI projects
- Easy access to your ML models for lab scientists
- Centralized ML model management
- Automated ML experiment logging
- Efficient ML model deployment
- Intuitive analysis interface and visualizations for lab scientists
Trusted by industry leaders













Transform your research with seamless data integration and future-ready data science
Contact us for a personalized demo or consultation.