Discovery analysis
Tooling
Unlock the full potential of your discovery research with our platform’s intuitive user interface, designed for scientists of all backgrounds. Whether you’re working with NGS, Sanger, single B cells, or multiple technologies, our platform integrates all sequence and assay data seamlessly.
- Hybridoma
- Single B cell
- Display panning
Empower your discovery with intuitive,
cutting-edge software
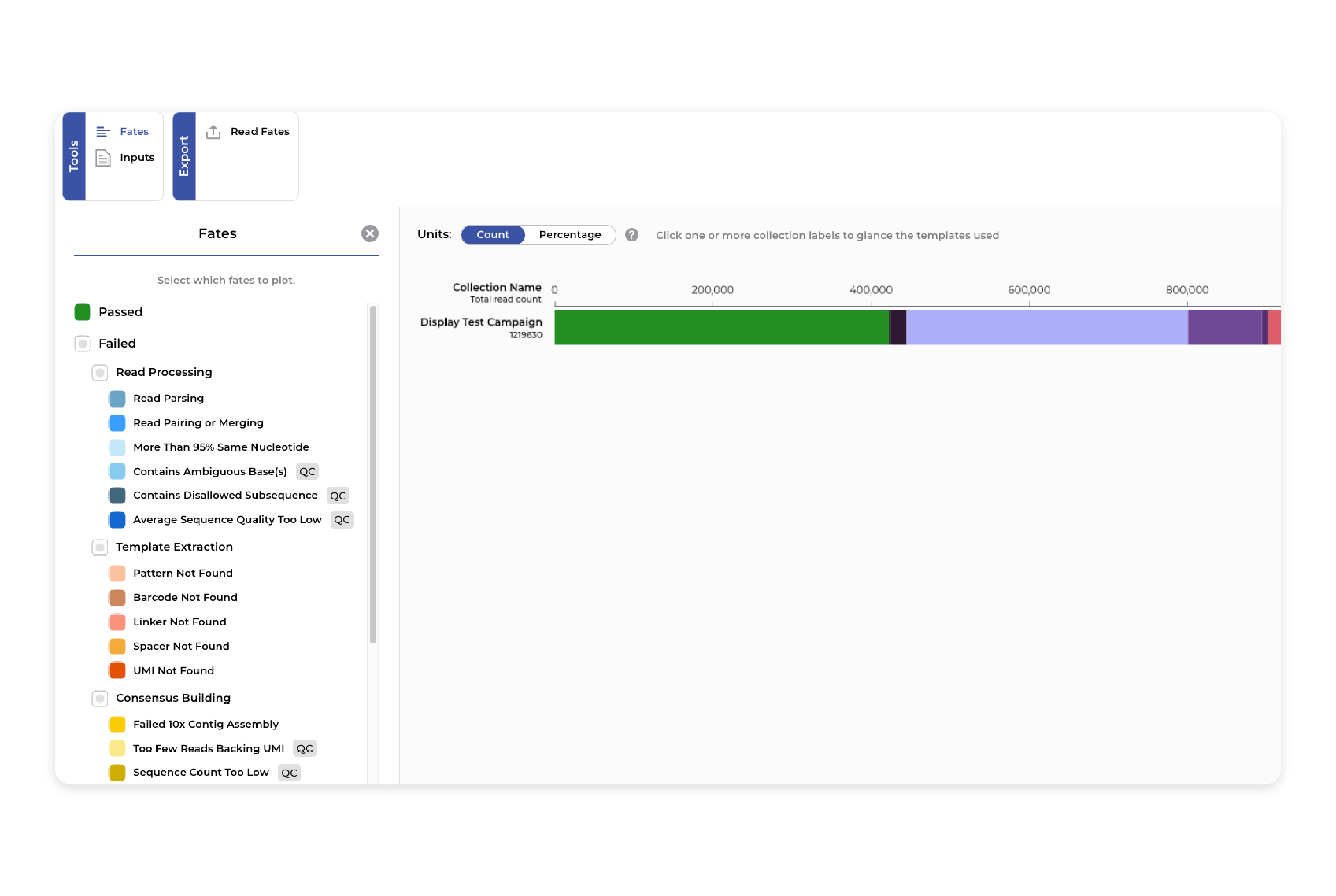
Quality control
Our platform supports quality control both before and after annotation. It provides FASTQ quality assessments with different interactive visualizations a Read Length Distribution and Average Base/Sample Quality. For Ab1 we have alignment-aided base edit to reduce spurious errors before annotation. Users have flexible settings for quality filters during processing and annotation and can inspect the impact in interactive read fate plots.
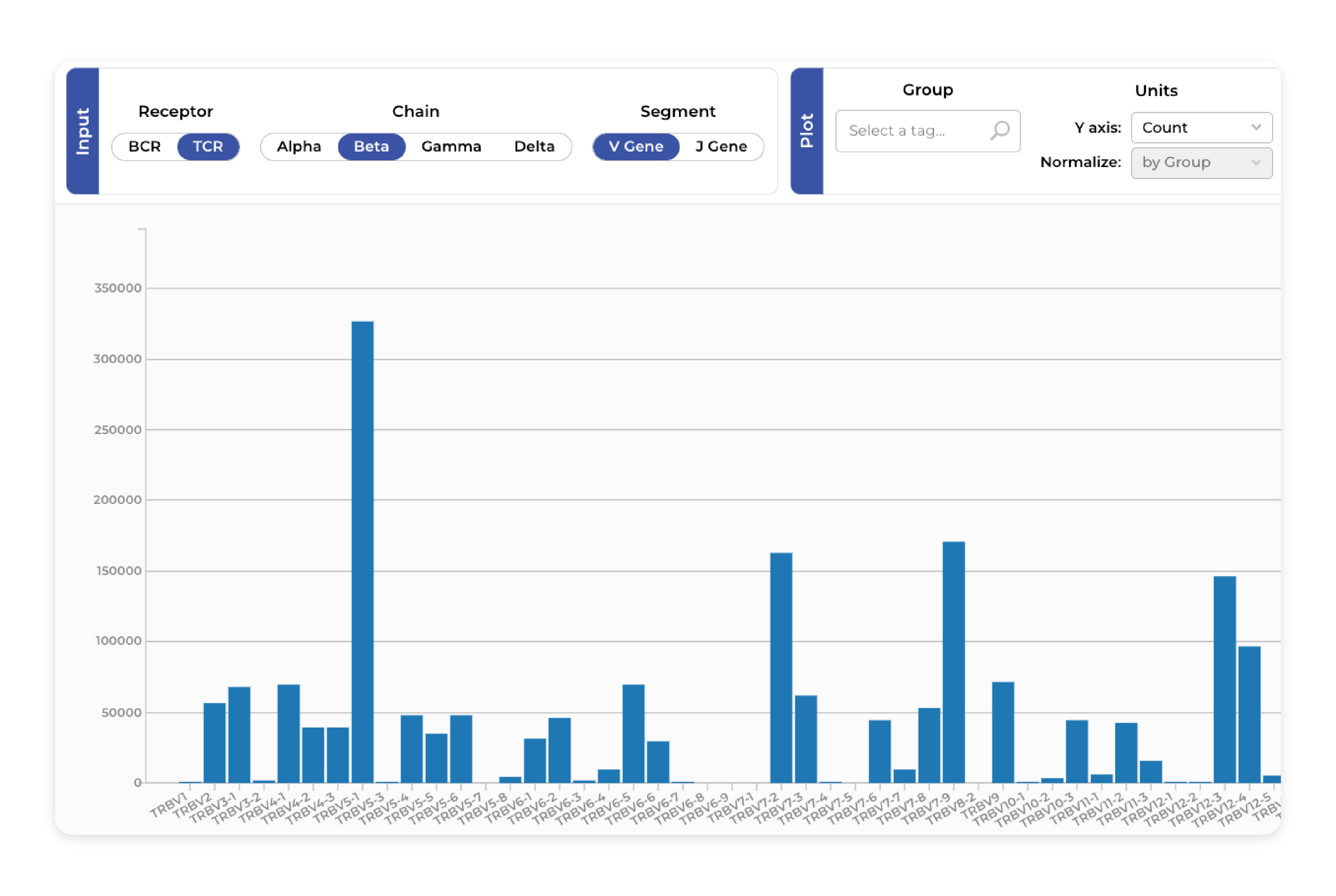
Repertoire overview
Users can easily generate intuitive visualizations that provide insight into the dynamics of the immune system response. It includes several analyses with interactive plots: V/J Gene Usage, Diversity Measures, and CDR3 Length Distribution.
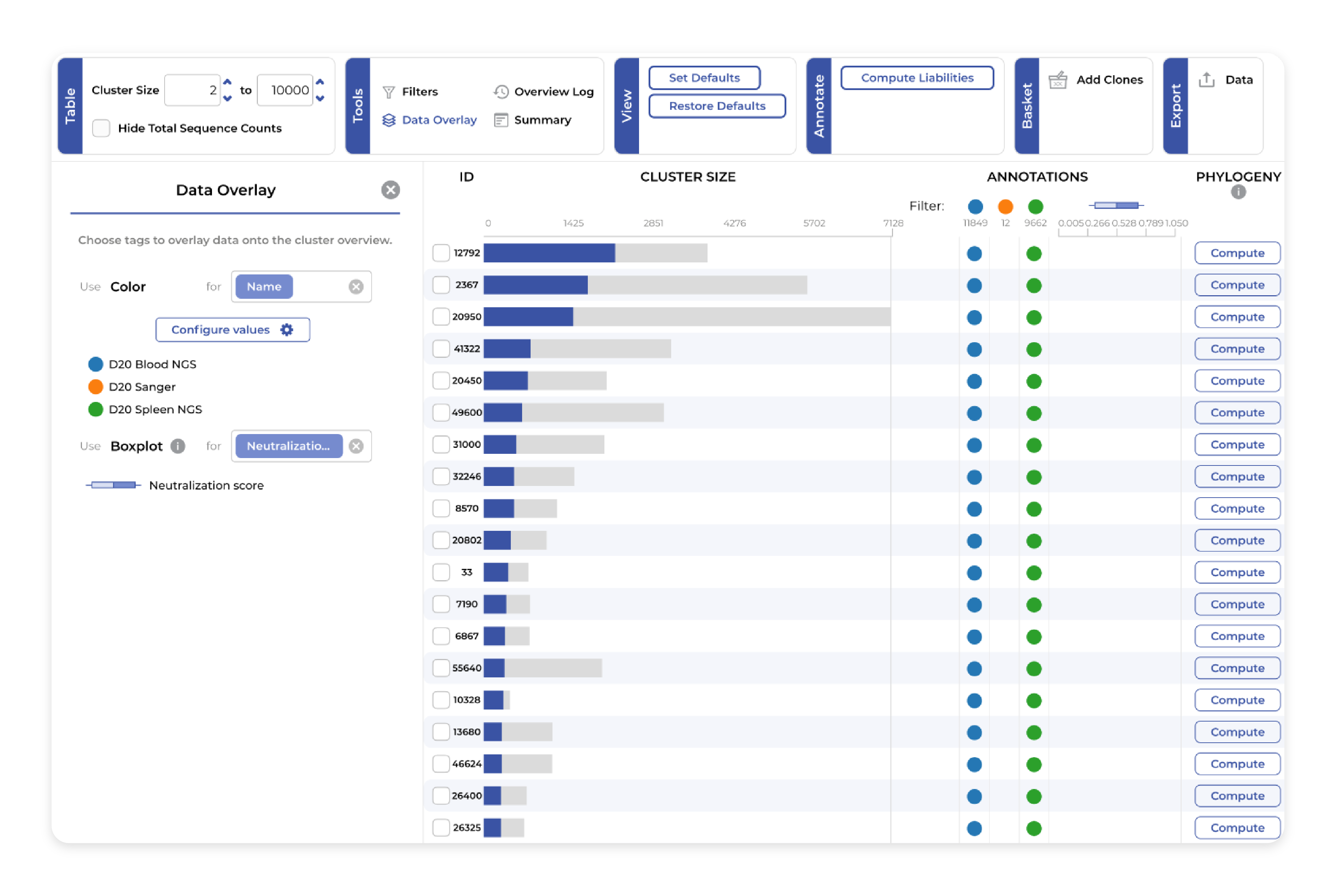
Clustering
Group clones and chains into clusters based on flexible, user-defined regions: whether CDR3, CDR123, VDJ, single or paired chains. Define these options and additional restrictions such as gene usage and CDR3 length in just a few clicks. Clustering samples from different sequencing technologies, campaigns, or rounds providing the ideal setup for important discovery tasks like candidate selection and hit expansion.
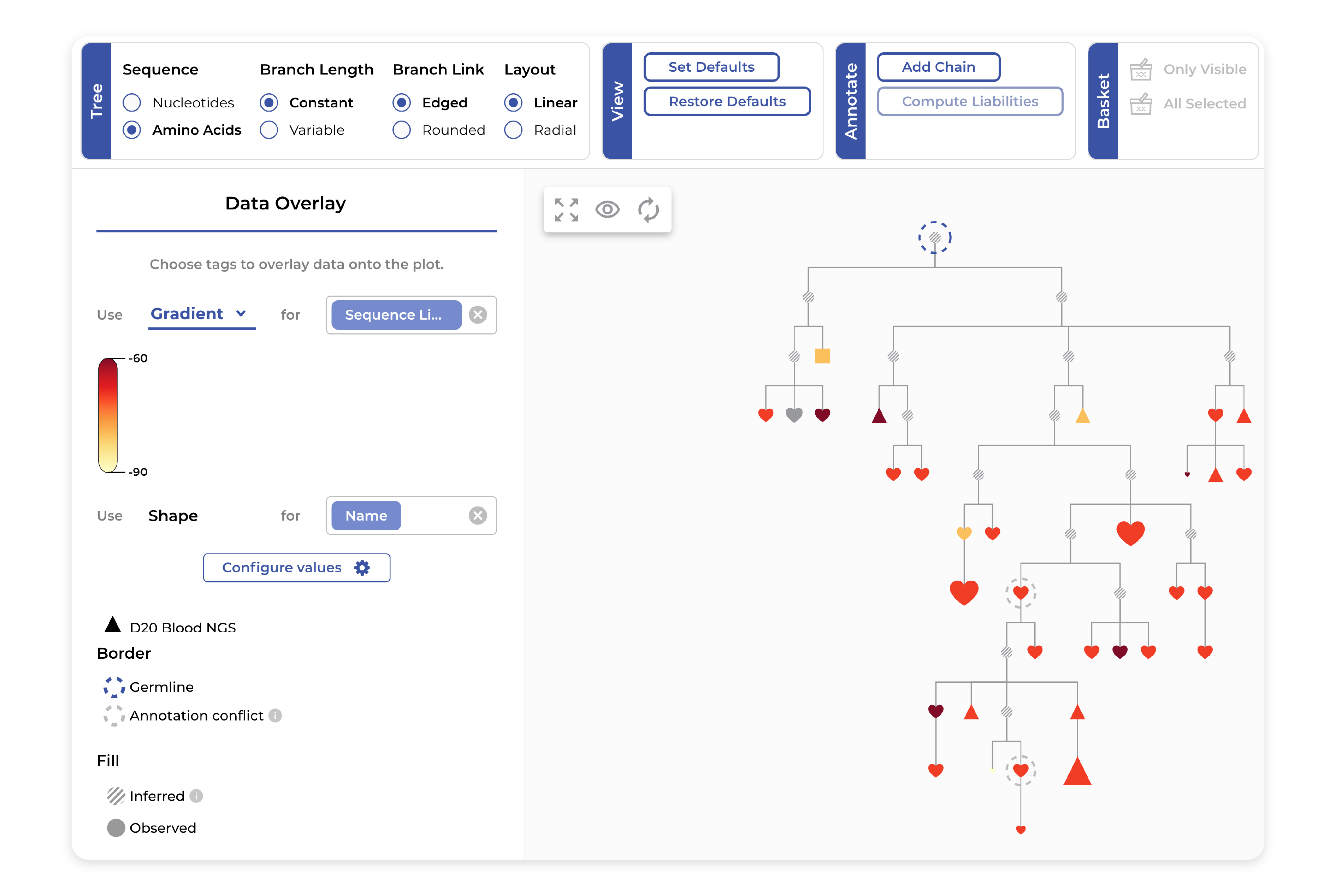
Phylogeny
Exploring clusters of interest and their phylogenetic evolution is simple with our interactive tree visualizations which link color, shape, and size to available experimental and AI-generated data. Aligning the sequences and seeing their associated metadata makes it easy to pick antibody candidates for follow-up analysis.
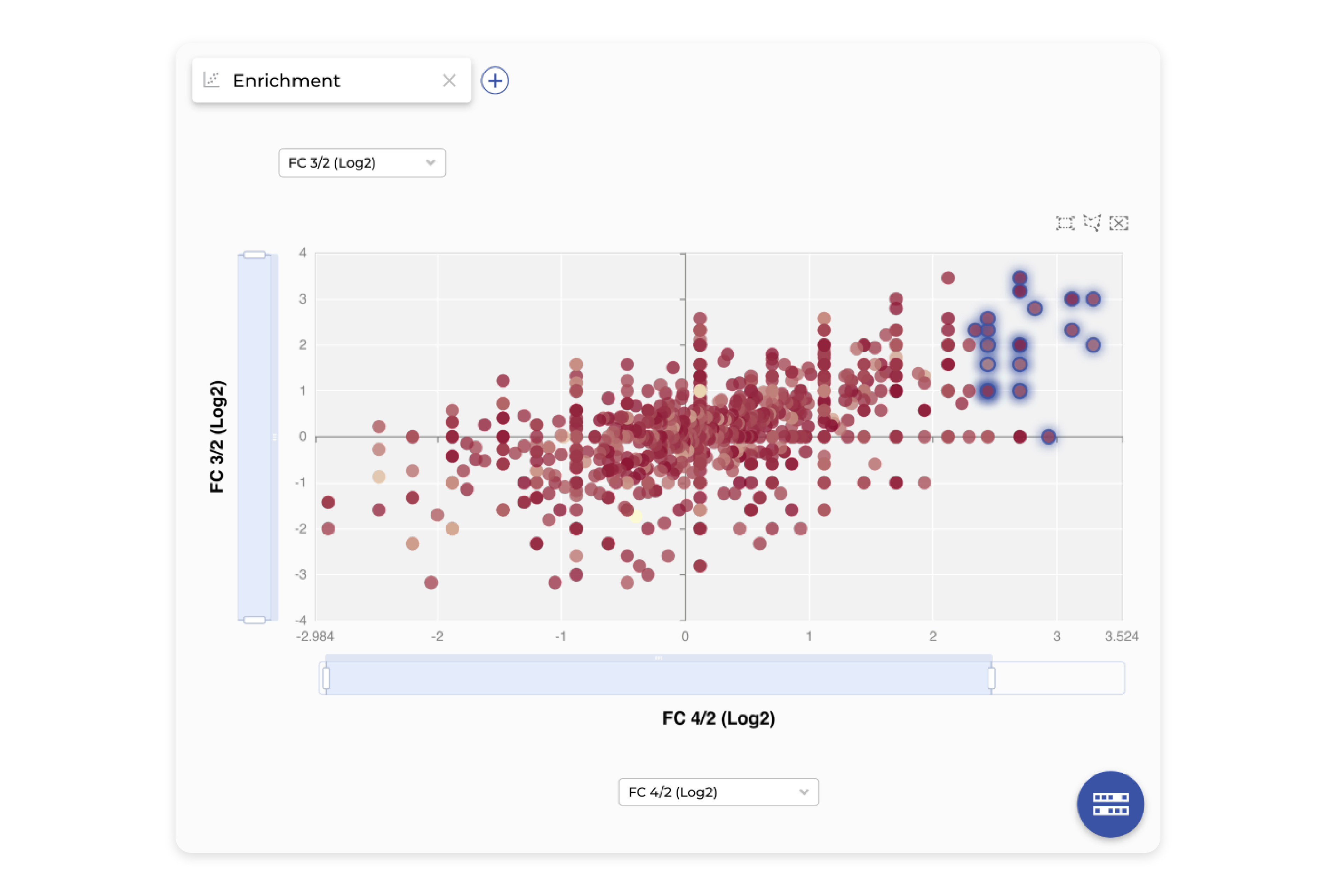
Enrichment
We provide a fast, flexible way to discover a diverse set of highly enriched candidates from display screening. Freely join, intersect, and subtract datasets to match the analysis to any panning campaign design, no matter the complexity. Explore the results through interactive tables and graphs, where any assay or predicted data can be integrated and combined with powerful filtering and sorting options to select the most promising leads.
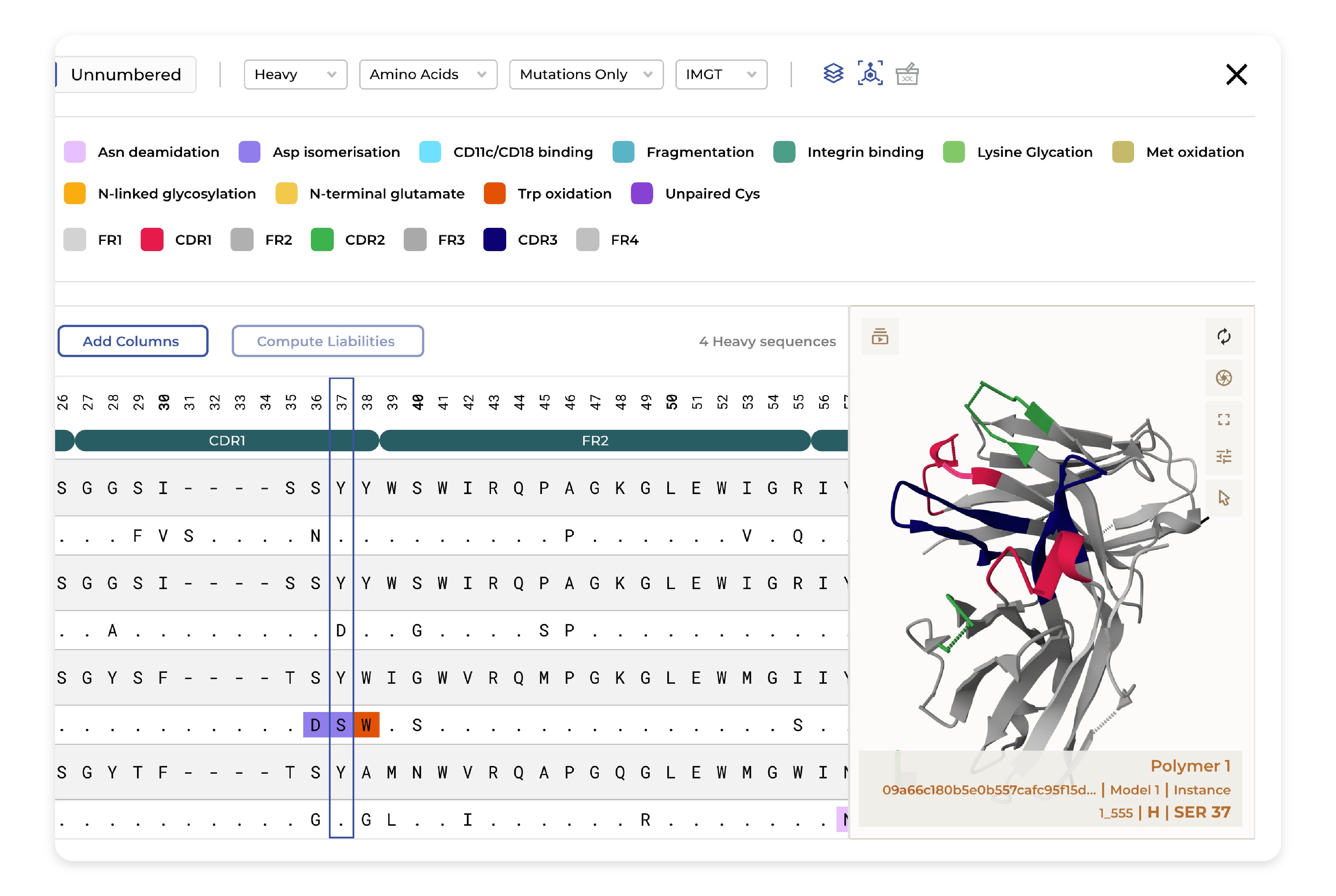
Developability
Strong binding is not sufficient for a developable biologic drug; this is why we enable high-throughput structural modeling of biologics to accurately predict and annotate exposed liabilities. Using the 3D model predicted our platform helps assess the developability of candidates by comparing the characteristics of your antibodies to those of clinically approved antibodies.
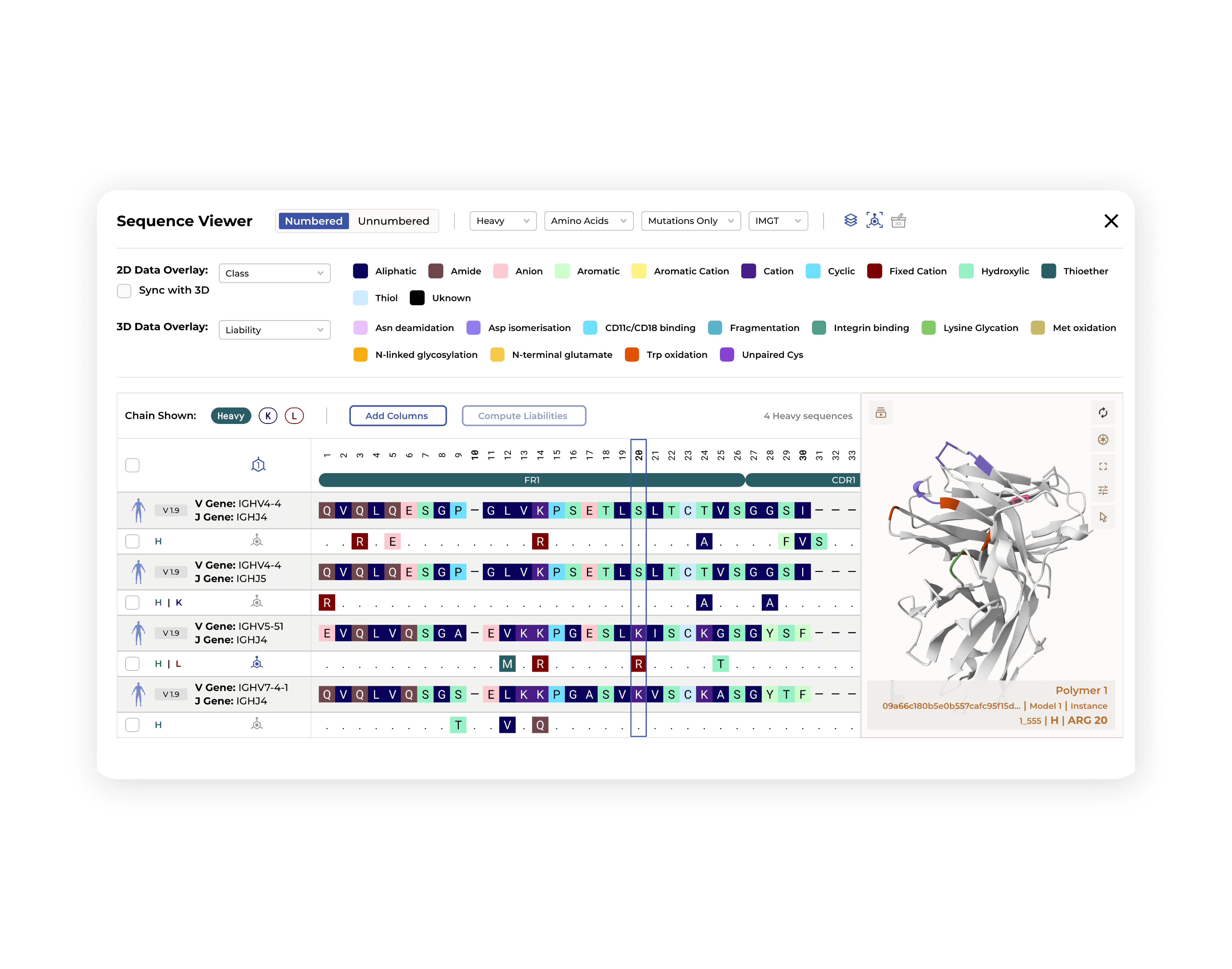
Data-driven candidate selection with rich visualizations
Lead selection involves evaluating an array of characteristics and molecular properties, and our platform empowers you to do this with ease. Associate any assay or experimental data, including in-silico predictions, directly with your clones and overlay them through information-rich visualizations. Streamline your workflow towards functionality-first selection, ensuring every candidate is assessed with all relevant information at your fingertips. Leave guesswork behind—embrace data-driven discovery, backed by actionable insights, and aligned with your ultimate therapeutic goals.
Join the future of clone annotation
Experience and accuracy in clone annotation
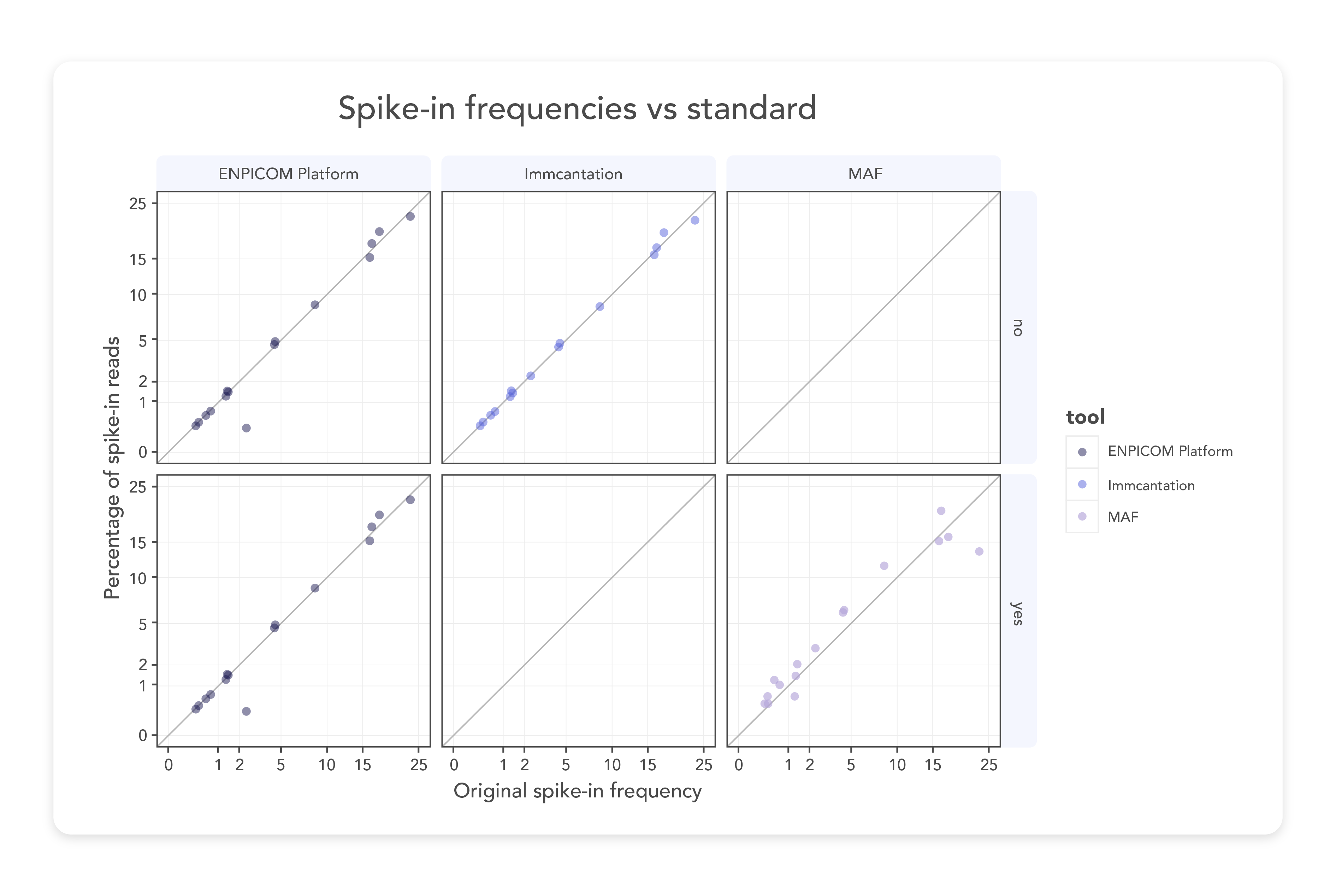
Our clonal annotation tool is the result of years of expertise in repertoire sequencing data annotation. The ENPICOM team has carefully optimized it to handle every step of the process with unmatched accuracy. From read-pair merging and quality filtering to UMI processing and chain-pair linking via barcodes, the ENPICOM Platform annotates BCR, TCR clones, and other molecules with exceptional accuracy. The sensitivity of these annotations has been rigorously benchmarked against spike-ins across diverse datasets, demonstrating its reliability and effectiveness.
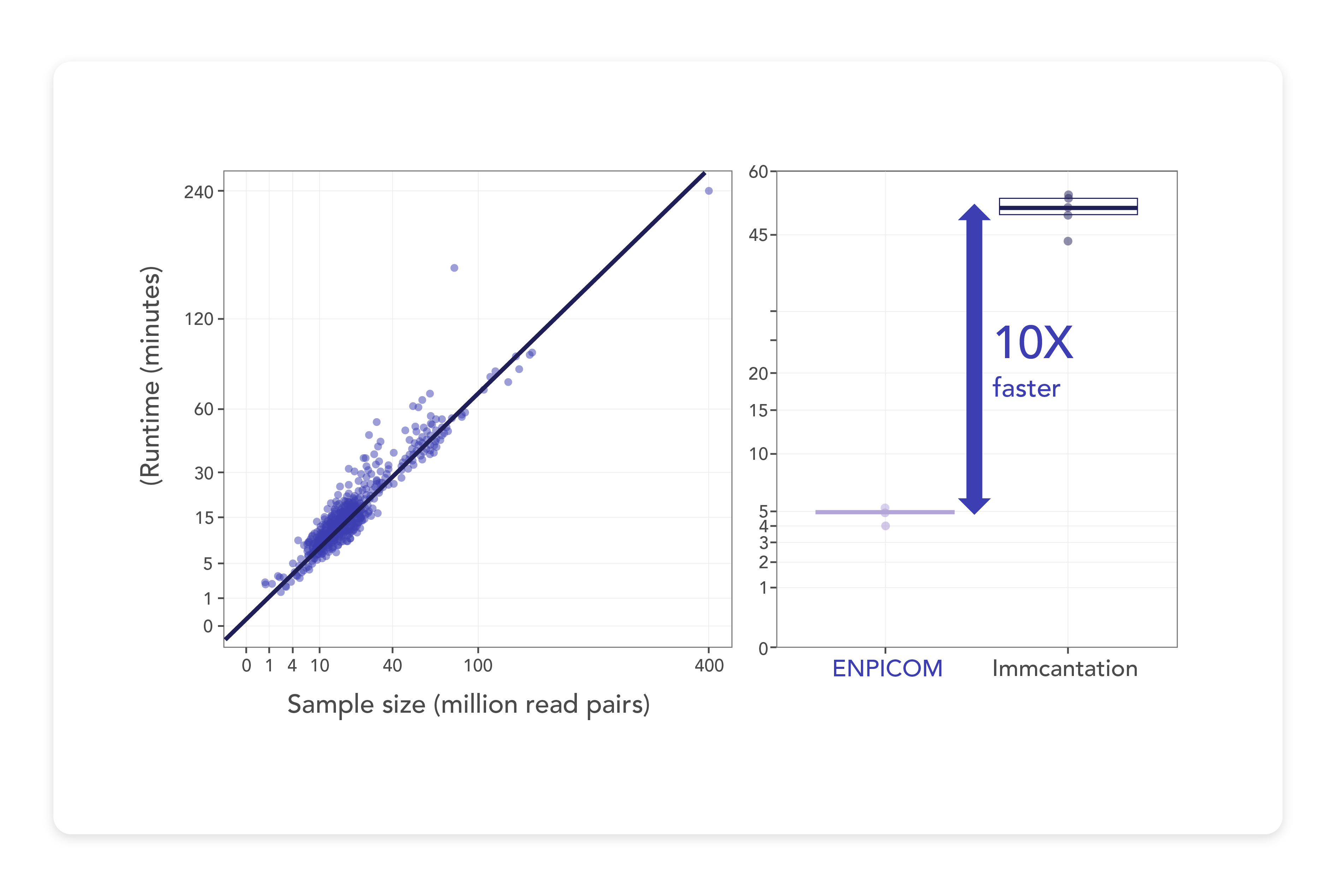
Blazing fast performance, built for scale
Purpose-built to handle datasets of any size, the ENPICOM Platform delivers blazing fast performance. Whether handling Sanger Ab1 files or large-scale NovaSeq FASTQs, ENPICOM enables seamless annotation and integration of all data within a unified platform. Capable of processing thousands of samples in parallel at speeds exceeding 1 million reads per minute, it effortlessly annotates billions of reads within just hours. This makes the ENPICOM Platform the fastest annotation tool available, boasting ten times the processing speed of other public solutions, and significantly reducing both the waiting time and costs associated with large sequencing datasets.
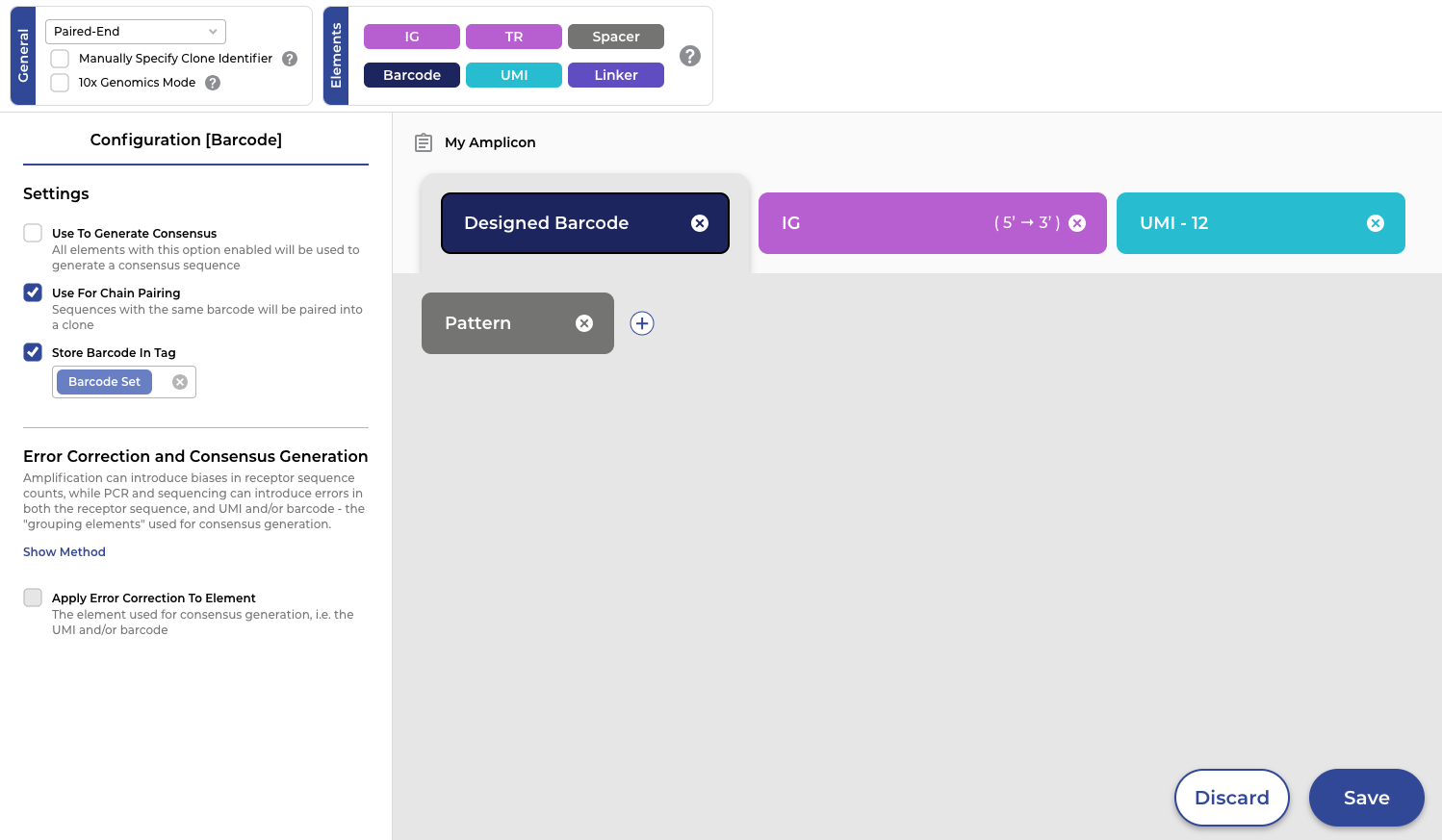
Fine control with ease of use
Even with its powerful performance, the ENPICOM Platform does not compromise on flexibility and user control. It offers extensive options for any amplicon/read configuration and quality control filtering through an intuitive interface. Whether processing single-chain VHH, scFv, or short-read 10x Genomics data, the configuration is straightforward with a simple drag-and-drop template. Scientists can also readily filter the data by read quality, sequence count, or UMI, restrict clones to those with both chains, and much more within a QC template. These templates streamline the setup process, ensuring consistency with SOPs and enhancing reproducibility across your projects.
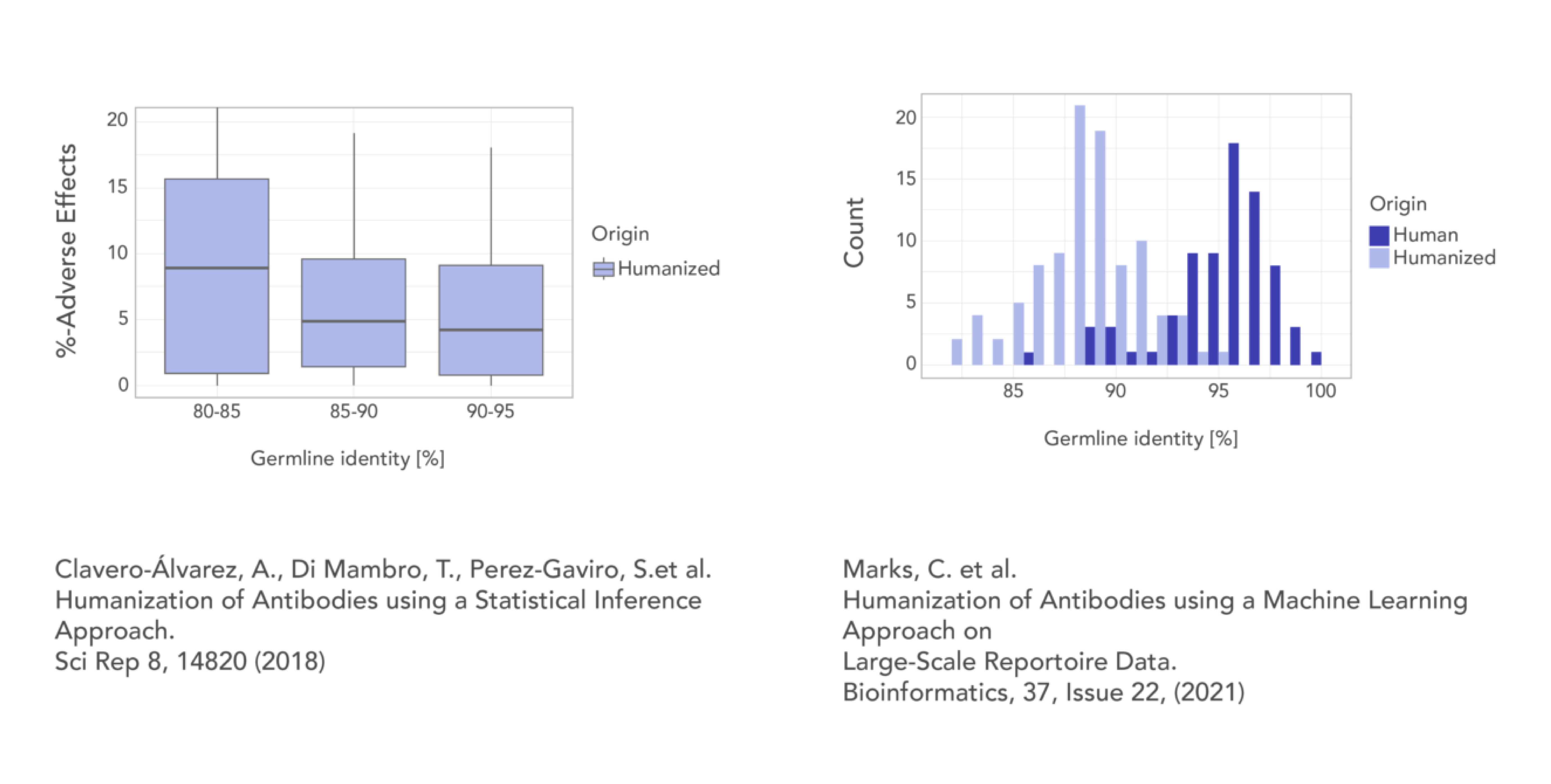
ML-powered humanization
Our antibody protein language model humanizes antibody candidates while maintaining high germline identity and superior binding affinity, outperforming industry standards. It supports humanization from any species, requiring only 3-5 variants to achieve clinically relevant humanness levels (>85% and >90% germline identity) while retaining binding affinity.
Leaders in scientific research choose ENPICOM













Transform your research with seamless data integration and future-ready data science
Contact us for a personalized demo today.