Antibody and TCR discovery rely on an array of sequencing technologies: Sanger, single B-/T-cell, and NGS are often used separately or in combination. While generation of sequence data has gotten significantly cheaper and easier, annotating immune receptors correctly and in a scalable manner has not. Powerful, specialized bioinformatics pipelines are required to accurately interpret large-scale repertoire data and extract biologically and clinically relevant insights. Let’s dive into key challenges of correct receptor identification and quantification and why this fundamental step in the analysis of immune repertoires requires tailor-made methods.
Challenges in accurate sequencing data annotation
One of the complexities of the annotation of B and T cell receptors is that they are generated by somatic recombination of gene segments, which results in millions of possible combinations of unique sequence features with variable lengths. Their analysis requires specialized algorithms that can handle the non-templated sequence elements, low signal-to-noise ratio, and the high variability found in immune repertoire data.
In therapeutics discovery, one of the major challenges is the diversity of the samples and formats used in a single campaign. Sanger, single-cell, and NGS sequencing each have their own unique strengths, requirements, and processing needs. Sanger sequencing is not very prone to errors but has limited quality information and throughput. NGS sequencing generates massive datasets that require UMI identification and error correction to ensure accurate results. Single-cell sequencing can track chain pairing, often using barcodes or indices. Apart from sequencing approaches, there is also ample variety in (antibody) construct formats like scFvs and FAbs, where the two chains are cloned into the same construct. All these different formats and sequencing methods require different processing and annotation strategies.
Importance of data preprocessing
The sequencing technology used to read these sequences each introduce errors, biases, and artifacts, which need to be identified and corrected. For example, NGS can have quite significant error rates and the library preparation process can lead to amplification biases, especially if multiplex amplification is used [1]. Template switch is not without issues and can lead to longer amplicons and lower quality of merged reads [2]. While NGS has opened the door to the deep interrogation of the immune repertoires, one of the key challenges is still the efficient and accurate analysis of the huge amounts of data that are generated. These datasets are becoming even larger with further advances in sequencing technologies, such as Illumina’s NovaSeq and NextSeq. This makes it increasingly important to have accurate and reliable tools that can scale to extremely large amounts of data.
Due to the diverse and complex nature of immune repertoires, and, moreover, the variety in sequencing protocols, different configurations are required to properly (pre)process the data. Indices, UMIs, and barcodes, when properly integrated, can help remove artifacts, minimize biases, and correct errors during data preprocessing. This is a critical step because the quality of the preprocessed data determines the accuracy and reliability of the subsequent clone annotation step.
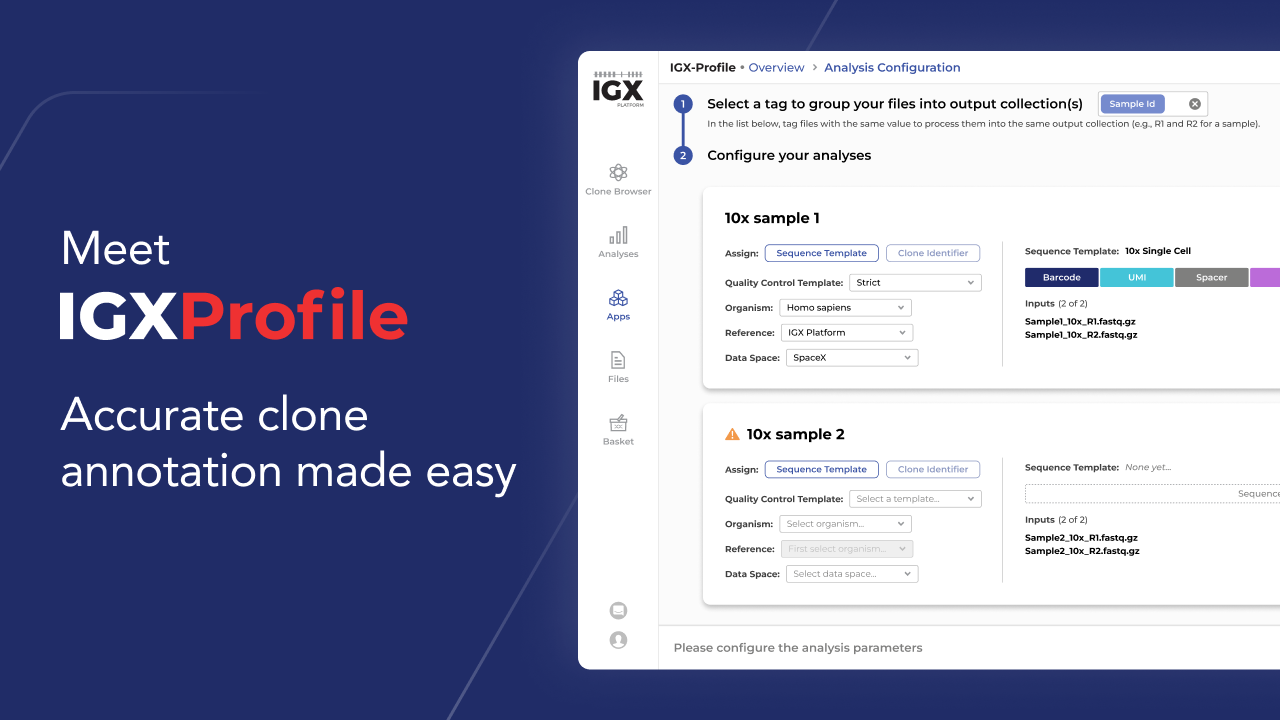
IGX-Profile: Accurate clone annotation made easy
At ENPICOM, we understand these challenges and have developed a solution to address them. Our new immune repertoire processing App, IGX-Profile, is specifically designed to handle any format and amplicon structure, including UMIs, barcodes, spacers, and linkers. It supports error correction and consensus generation, that considers base quality and frequency of occurrence in its proprietary UMI error correction. Extensive QC options allow you to finetune your quality standards and discard low-evidence receptors.
IGX-Profile processes any type of single or paired chain data, using either (1) header or filename information (often used in Sanger sequencing), (2) linkers between chains (e.g. scFv), or (3) barcodes or indices (like 10x Genomics or plate-based single-cell sequencing). Moreover, it is built with a scalability-first approach, processing anywhere from one to one hundred million billion sequences.
Unlock next-generation Rep-Seq data profiling
Whether you are developing new therapeutic antibodies or studying immune responses in the clinic, with IGX-Profile you can be sure that your clones will be accurately annotated and ready to be discovered in one of our downstream analysis Apps.
Would you like to experience IGX Platform’s new data processing capabilities firsthand? Schedule a personalized demo today.
References:
- Barennes, P., Quiniou, V., Shugay, M. et al. Benchmarking of T cell receptor repertoire profiling methods reveals large systematic biases. Nat Biotechnol 39, 236–245 (2021). https://doi.org/10.1038/s41587-020-0656-3
- Vázquez Bernat N, Corcoran M, Hardt U, Kaduk M, Phad GE, Martin M, Karlsson Hedestam GB. High-Quality Library Preparation for NGS-Based Immunoglobulin Germline Gene Inference and Repertoire Expression Analysis. Front Immunol. 2019 Apr 5;10:660. doi: 10.3389/fimmu.2019.00660. PMID: 31024532; PMCID: PMC6459949