The pharmaceutical industry has been trying to further increase the value out of their drug discovery campaigns to accelerate delivery to market and reduce research costs. Traditional hybridoma-based discovery approaches are suffering from low scalability and a saturated ecosystem, in which most ‘low-hanging fruit’ antibodies have already been discovered. The innovative single B-cell discovery platforms address these concerns, but the equipment and setup costs may be too high for many research groups to integrate it into their workflow. Display technologies offer alternative platforms that can improve scalability and research scope at a lower cost compared to single B-cell technologies. In this blog, we will discuss display technologies, enrichment analysis, and how the IGX Platform can help you make the best out of your display-based antibody discovery campaigns.
Display technologies offer high-throughput discovery workflows with control over the initial antibody repertoire and the candidate selection process. In display technologies, Fab, scFv, VHH, or other antibody constructs are recombinantly expressed in display organisms (phage, yeast, mammalian cell, or cell-free), which are then repeatedly screened for binding against the antigen. The weak or non-binders are washed away during the screening process while the strong binders remain bound to the antigen and are amplified before the next selection round. The iteration of these screening rounds, also known as ‘panning rounds’, aim to enrich the library for antibodies specific for the target. Typically, three to four rounds of panning are performed before acquiring promising leads.
Diverse types of libraries can be used in display technologies, such as naïve, immunized, and even synthetic and semi-synthetic antibody repertoire libraries. These antibody libraries can reach repertoire sizes of 1010-11 unique clones [1], which goes at least an order of magnitude above the estimated in vivo circulating B cell diversity of 107-9 [2]. This means that, if the screening process is performed accurately, there is the potential of discovering a large number of binders against most targets. However, relying solely on in vitro selection can yield a limited set of candidates, since the screening is affected by multiple factors including the initial clonal frequency, the screening conditions, and other experimental parameters. These factors are not always easily controlled, which can lead to biases and cause difficulties with the selection of antibodies with desirable binding properties for therapeutic use. If the display campaign does not yield a diverse set of candidates, researchers may need to alter the experimental conditions and repeat the screening process, which further increases the cost of research. On the other hand, if the display campaign has a lot of promising candidates, it becomes infeasible to experimentally characterize each candidate, which might lead to selecting a subset with limited information.
Next-generation sequencing (NGS) can be used to characterize the panning rounds output and mitigate these issues. If a selection round was too stringent, the antibodies present in previous rounds can be still tested. If it was too relaxed, then metrics like the initial clonal count of the fold-change between rounds can help decide which candidates have most potential. If the round is sequenced at sufficient depth, the fold-change between panning rounds can be linked to the enrichment, which can correlate with the binding affinity in certain panning conditions [3]. This correlation though is currently the source of debate since recent studies have failed to find it [4, 5]. Even without utilizing enrichment in the selection criteria, NGS also provides the initial clonal count that can signal library biases and help researchers figure out the clones that survive the panning rounds not because of their superior binding but due to their abundant numbers. High “on-target” count is not the only characteristic that defines a good therapeutic antibody, as spurious binding of irrelevant antigens is a frequent occurrence. Display technologies have been shown to generate antibodies prone to specificity and developability issues [6]. Phage display in particular has been shown to have a higher risk of selecting non-specific antibodies due to the higher levels of aliphatic residue content in the generated antibodies compared to their animal counterparts [7].
Moreover, there might be a need to avoid binding to specific epitopes on the target antigen. Display technologies enable identification of the unwanted binders by including additional panning conditions on “counter-targets” as well as “irrelevant” antigens, offering a degree of control far beyond what is available in immunized animals. Panning rounds against irrelevant targets or counter targets can be extremely useful to discard un-specific or “sticky” antibodies present in the initial library. Generating multiple screening rounds outputs of various panning conditions (e.g., on-target, counter-target, irrelevant) increases the sensitivity and precision with which one can pick the best candidates.
With increasing panning conditions and rounds come additional datasets, which, in turn, increases the need for sequencing and the complexity of the data analysis and computational burden. Setting up an analysis including all panning conditions and rounds is technically challenging, due to the number of datasets, their scale, and the operations one must perform on them (i.e., join, intersect, subtract different rounds of panning). Finally, incorporating knowledge of previously discovered candidates, wet-lab assay data, and structural predictions is crucial to making fully informed decisions and obtaining a high-quality lead list.
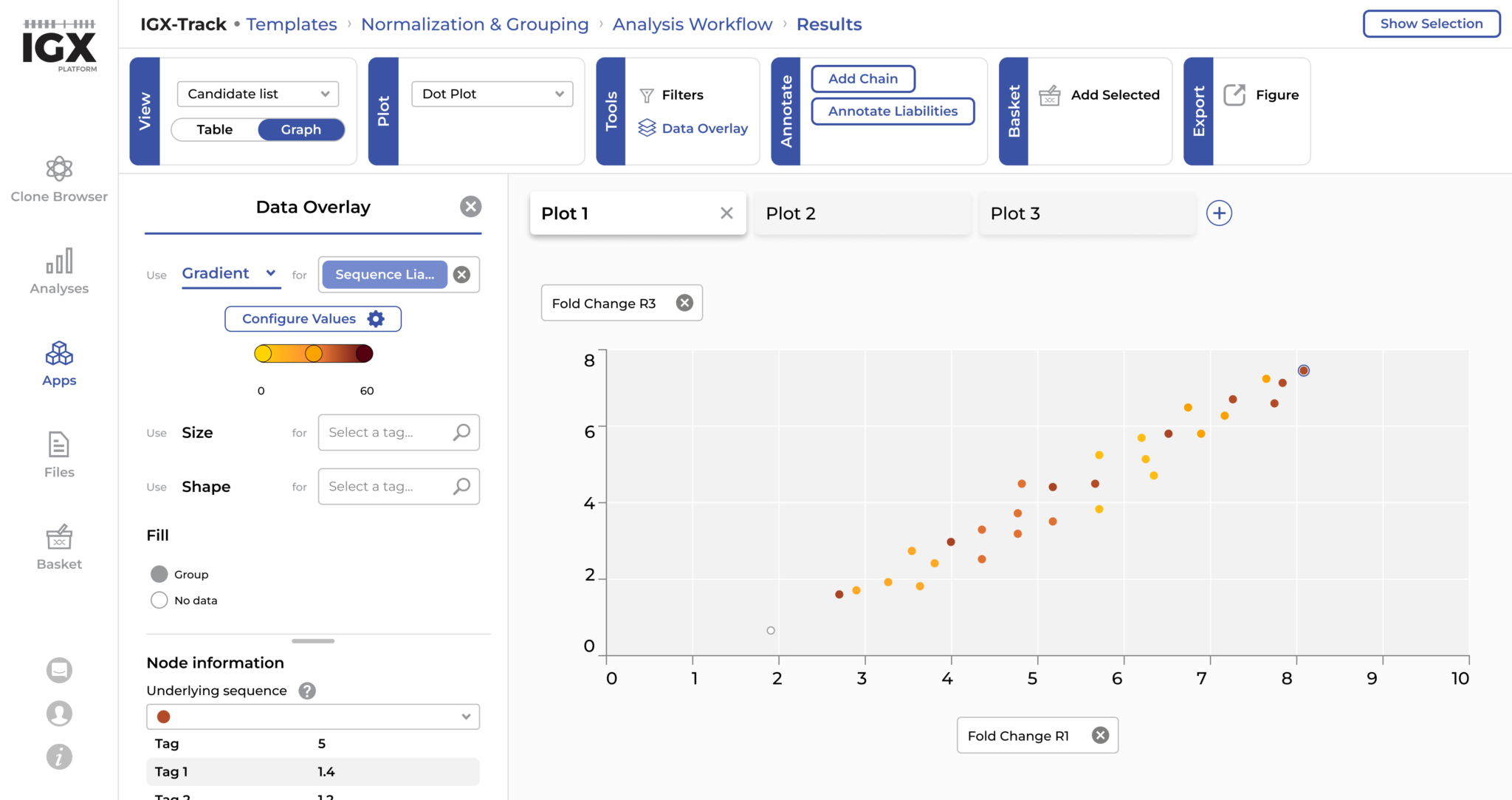
At ENPICOM, we have created IGX-Track to support the enrichment analysis of any display technology and harness the full potential of these platforms. IGX-Track provides a fast, flexible way to discover a diverse set of highly enriched candidates from display screening. By allowing users to freely join, intersect and subtract datasets, its visual workflow builder supports the analysis of any panning campaign design, no matter the complexity. For example, antibodies binding irrelevant antigens in control panning conditions can be easily subtracted to increase candidate selection precision. Results can be explored through interactive tables and graphs, where any metadata or assay data can be integrated and combined with powerful filtering and sorting options to select the most promising antibody candidates.
Moreover, IGX-Track is integrated into our IGX Platform which allows us to leverage the 3D-model-based liability predictions during the candidate selection and utilize the unique Basket environment to collect all the promising clones. Together with the IGX Platform, NGS sequencing allows researchers to fully capitalize on the library diversity provided by display technology. It enables rational selection of enriched clones with increased diversity and quality of selected leads.
The IGX Platform enables you to select the most promising antibody candidates from your display panning experiment by:
- Tracking clones across panning rounds to identify highly enriched candidates through interactive, user-defined visualizations.
- Allowing users to freely join, intersect, and subtract your datasets to match your panning campaign design.
- Avoiding picking sub-optimal candidates by overlaying accurate liability predictions, assay data, and more onto enrichment graphs.
Contact us for a demo today!
References
- Almagro, J. C., Pedraza-Escalona, M., Arrieta, H. I., & Pérez-Tapia, S. M. (2019). Phage Display Libraries for Antibody Therapeutic Discovery and Development. Antibodies (Basel, Switzerland), 8(3), 44. https://doi.org/10.3390/antib8030044
- Rees A. R. (2020). Understanding the human antibody repertoire. mAbs, 12(1), 1729683. https://doi.org/10.1080/19420862.2020.1729683
- Yang, W., Yoon, A., Lee, S. et al. Next-generation sequencing enables the discovery of more diverse positive clones from a phage-displayed antibody library. Exp Mol Med 49, e308 (2017). https://doi.org/10.1038/emm.2017.22
- Makowski, E. K., Kinnunen, P. C., Huang, J., Wu, L., Smith, M. D., Wang, T., Desai, A. A., Streu, C. N., Zhang, Y., Zupancic, J. M., Schardt, J. S., Linderman, J. J., & Tessier, P. M. (2022). Co-optimization of therapeutic antibody affinity and specificity using machine learning models that generalize to novel mutational space. Nature communications, 13(1), 3788. https://doi.org/10.1038/s41467-022-31457-3
- Mason, D. M., Friedensohn, S., Weber, C. R., Jordi, C., Wagner, B., Meng, S. M., Ehling, R. A., Bonati, L., Dahinden, J., Gainza, P., Correia, B. E., & Reddy, S. T. (2021). Optimization of therapeutic antibodies by predicting antigen specificity from antibody sequence via deep learning. Nature biomedical engineering, 5(6), 600–612. https://doi.org/10.1038/s41551-021-00699-9
- Jain, T., Sun, T., Durand, S., Hall, A., Houston, N. R., Nett, J. H., Sharkey, B., Bobrowicz, B., Caffry, I., Yu, Y., Cao, Y., Lynaugh, H., Brown, M., Baruah, H., Gray, L. T., Krauland, E. M., Xu, Y., Vásquez, M., & Wittrup, K. D. (2017). Biophysical properties of the clinical-stage antibody landscape. Proceedings of the National Academy of Sciences of the United States of America, 114(5), 944–949. https://doi.org/10.1073/pnas.1616408114
- Kaleli, N. E., Karadag, M., & Kalyoncu, S. (2019). Phage display derived therapeutic antibodies have enriched aliphatic content: Insights for developability issues. Proteins, 87(7), 607–618. https://doi.org/10.1002/prot.25685