Data foundation
The backbone of AI in biologics research
AI in biologics discovery and engineering is only as powerful as the data it learns from. Fragmented, inconsistent, and inaccessible data stall AI adoption and slow innovation. To fully leverage AI, annotated data must be stored in high-performance databases that enable rapid retrieval and seamless access to large-scale datasets. Findable, Accessible, Interoperable, and Reusable (FAIR) data drives scientific innovation by making information searchable, ready for advanced analytics, and able to flow freely across systems, tools, and teams.
The ENPICOM Platform is purpose-built to meet these demands. Designed for biologics, its data foundation integrates seamlessly into any ecosystem or federated data warehouse, eliminating silos. Powered by a proprietary high-performance architecture, it delivers instant access to all your data, accelerating discovery and AI-driven insights.
Built for biologics
The ENPICOM Platform is engineered to process large-scale biologics data with speed and accuracy. State-of-the-art algorithms and tools help you make full use of your sequencing and assay data to identify high-quality, developable candidates with greater efficiency.
Unmatched performance
Powered by a proprietary high-performance data architecture, the ENPICOM data foundation enables instant access to vast datasets. Query hundreds of millions of sequences in seconds, accelerating research and making the most of all your historical data.
Seamless integration
Eliminate data silos and fragmented systems. Our data foundation integrates seamlessly with existing systems and federated data warehouses, creating a single source of truth, FAIR by design, accessible across teams, and optimized for advanced analytics and AI applications.
Accelerate insights with high-performance data infrastructure
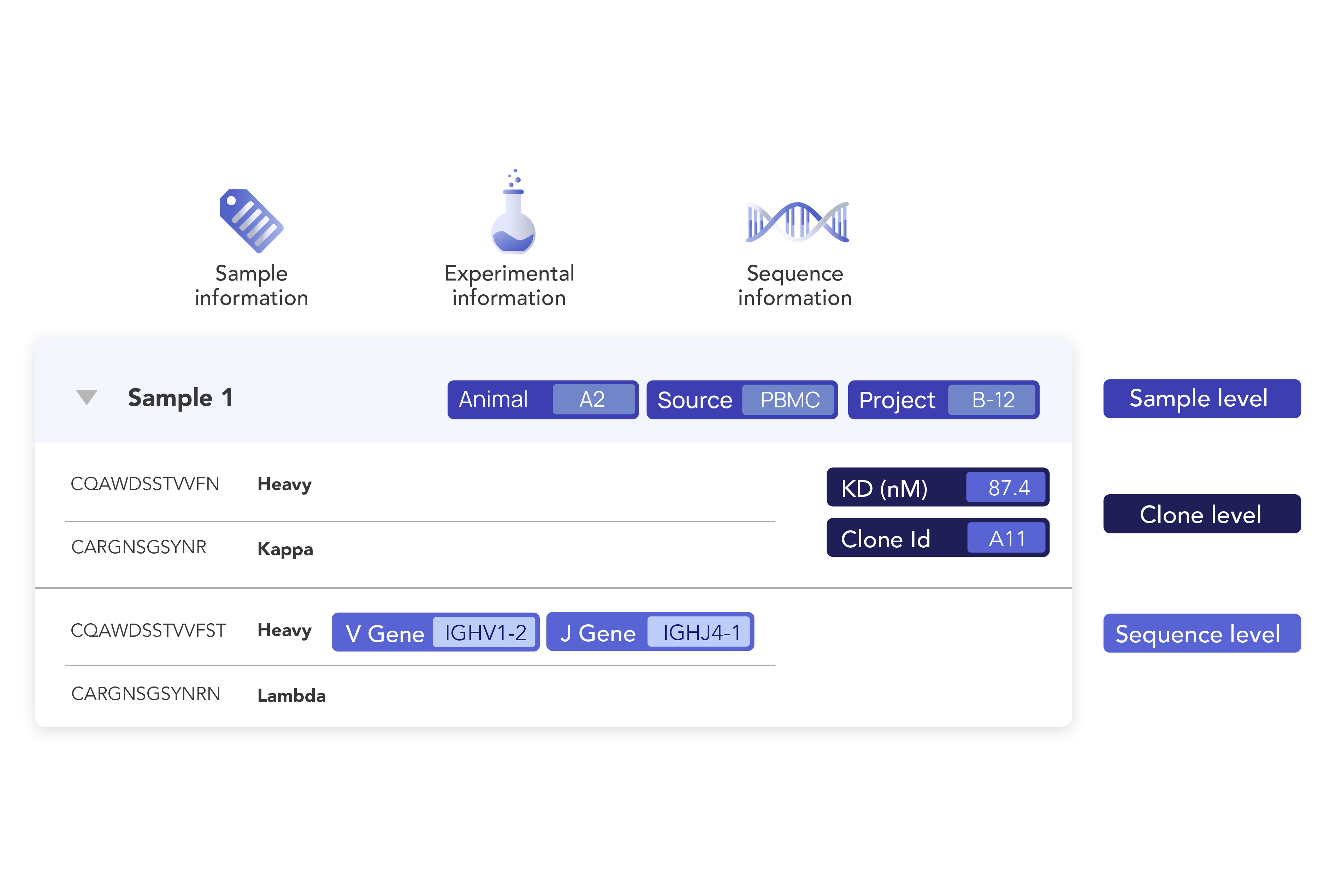
Effective data management is crucial to discovery analyses including diverse experimental metadata and/or large sequencing datasets, and the ENPICOM Platform excels in this domain. All annotated data is stored in a rapid, scalable database that ensures effortless access and extraction for analyses. This database houses sequencing data and enables seamless association of any assay or in-silico generated data with individual clones, allowing you to integrate these in your analysis to find the best leads.
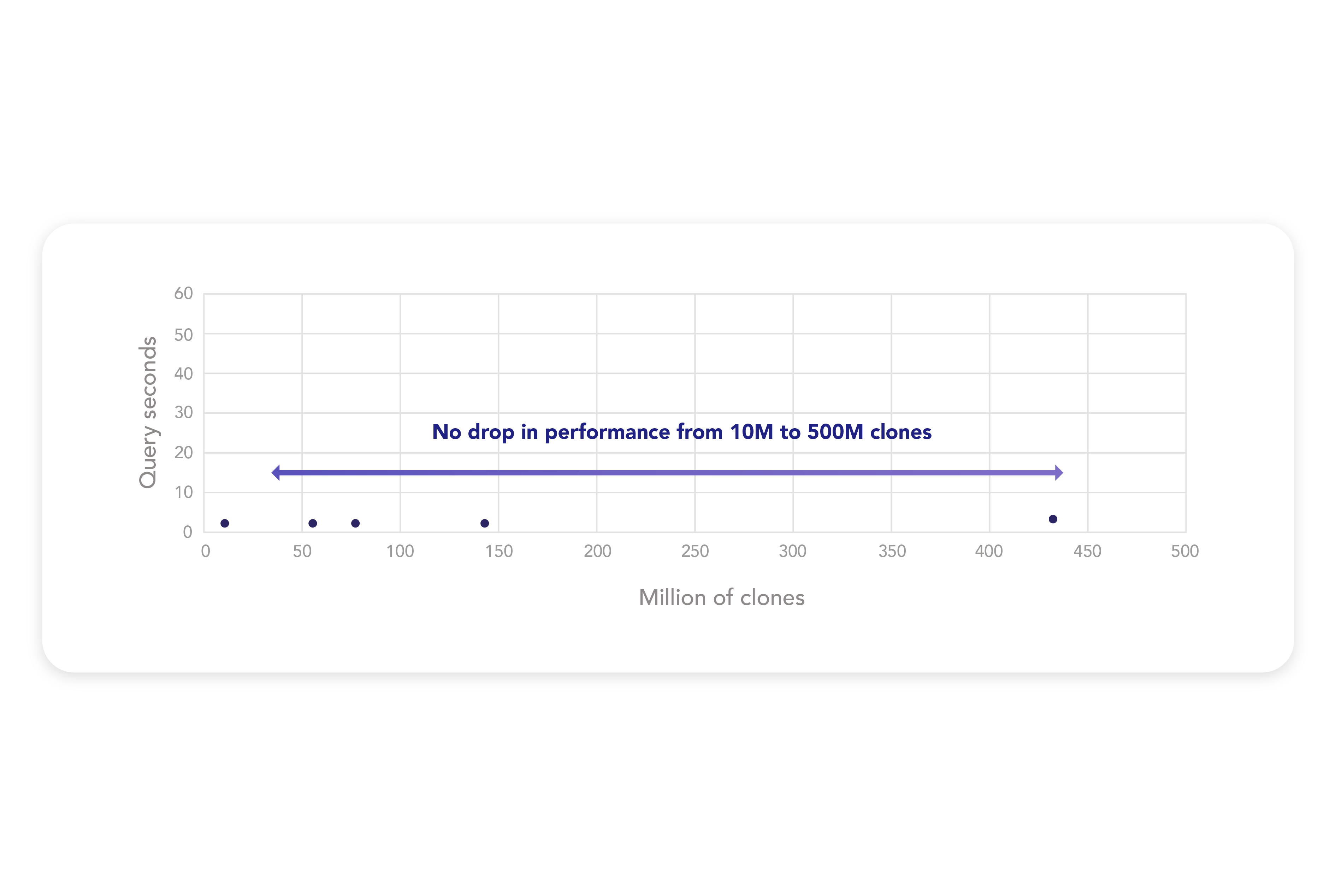
At the core of this system is Amazon’s Redshift Massive Parallel Processing (MPP) architecture, combined with our proprietary high-throughput SQL engine. This powerful combination allows for blazing-fast queries, enabling you to search receptor motifs among hundreds of millions in mere seconds. The database is built to scale to petabyte-sized datasets, ensuring smooth data access and retrieval. Querying massive datasets remains seamless, with no drop in performance from 10M to 500M clones.
Make all your data count. Now and in the future.
Easily structure and consolidate all your biologics metadata and experimental results to ensure accuracy, organization-wide standardization, and readiness for analysis, visualization, and AI integration. With well-organized data, you can easily augment and correlate experimental results within and across projects. Uniquely, our platform instantly incorporates new data into all previously conducted analyses, no manual reprocessing needed. This real-time data integration capability keeps your teams working with the latest insights, accelerating decision-making and improving scientific outcomes compared to traditional approaches that create isolated data snapshots.
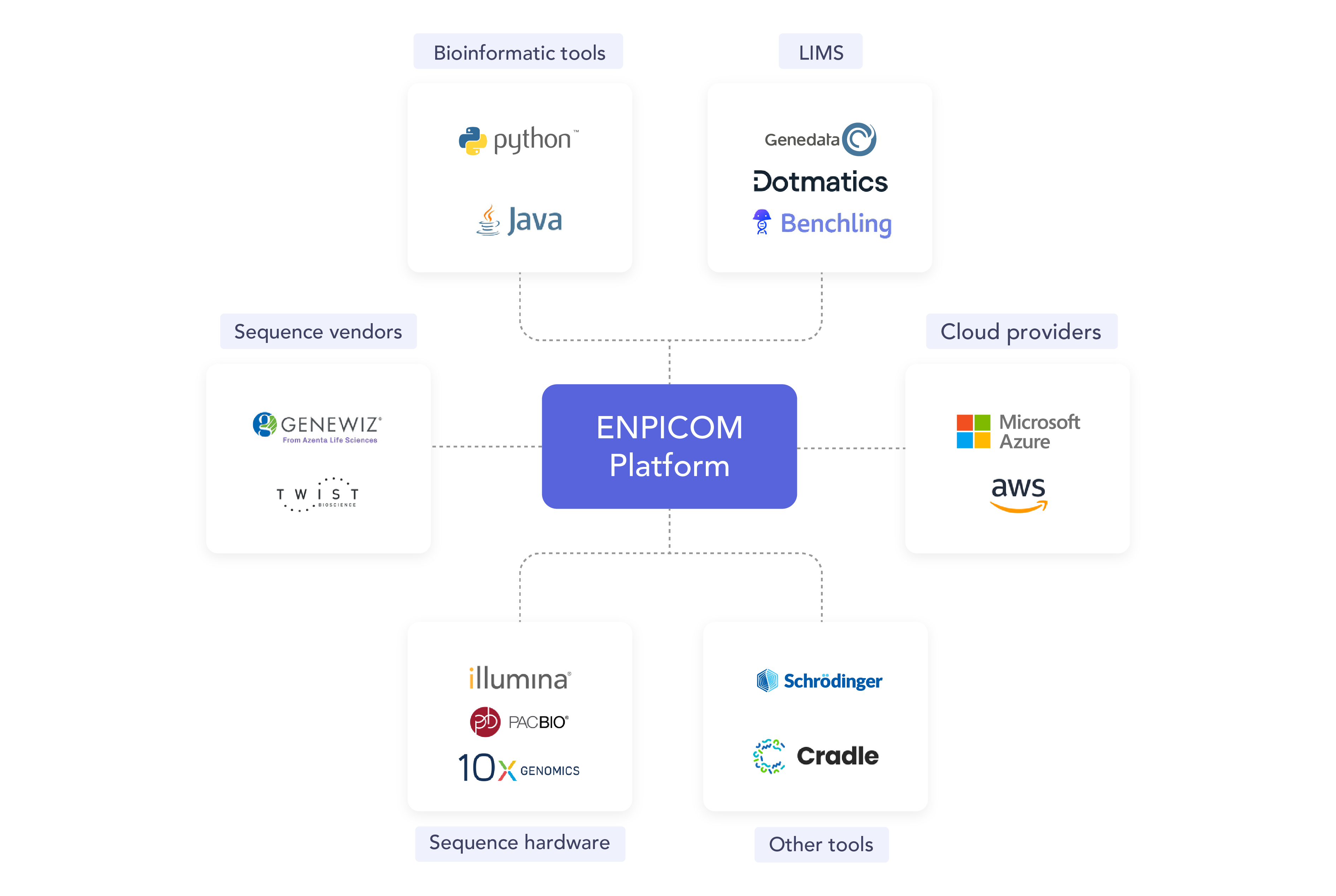
Collect and centralize your data
The ENPICOM Platform offers a centralized data architecture that eliminates fragmentation, providing a single source of truth for biologics research. It fully integrates with our partners’ ecosystems through a comprehensive API, enabling seamless connectivity with Laboratory Information Management Systems (LIMS), Electronic Laboratory Notebooks (ELNs), and various cloud storage solutions (like S3, Google Cloud, Azure) for automatic synchronization of experimental data and metadata. For developers, our user-friendly Python SDK simplifies the creation of custom workflows and integrations, empowering teams to tailor the platform to their specific project needs.
Extensive API and SDK
Drug development is a highly complex and multifaceted process, and with the increasing integration of AI into biologics research, seamless communication between various components has become more essential than ever. In this evolving landscape, different systems and tools must work together in harmony to contribute to the overall goal of accelerating discovery and development. This is why having a robust and stable API layer is critical.
Our robust API facilitates seamless data integration with external platforms, allowing users to easily pull data into the ENPICOM Platform and push results to other systems. Complementing the API is our user-friendly Python SDK, which simplifies the development process and empowers teams to customize workflows and integrations to meet specific project needs.
Data integration
Maintain a single source of truth for all meta-information by automatically synchronizing clone-related data, including assay results and liability predictions, with systems such as LIMS or ELN. You can also connect external data storage solutions, like Amazon S3, for automatic import or export of sequencing data.
Automation
Streamline your analysis process by creating automated workflows through an intuitive interface or via the API/SDK, while retaining the flexibility to explore results and conduct additional case-specific analyses.
Custom code
Execute proprietary algorithms to predict antibody properties, providing valuable outputs for scientists during candidate selection. You can also create custom visualizations or analyses with your data, or utilize tailored algorithms for sequence clustering, visualizing results directly within the ENPICOM Platform interface.
Data organization and AI
Leverage the scalable architecture and accessible interface of the ENPICOM Platform, along with the powerful API and SDK, to easily retrieve structured and labelled datasets for training your in-house machine learning models. This approach eliminates the time-consuming task of structuring datasets and simplifies querying for training purposes.
Industry leading security
Security of course is a top priority in our platform. We leverage AWS enterprise security, regularly conducting vulnerability scanning, penetration testing, and enforcing a zero-trust policy for database access. This ensures that your data is not only accessible but also protected to the highest standards.
- Security protocols
- ISO27001 compliance
- SSO integration
- 2-factor authentication (2FA)

Trusted by industry leaders













Transform your research with seamless data integration
and future-ready data science
Contact us for a demo