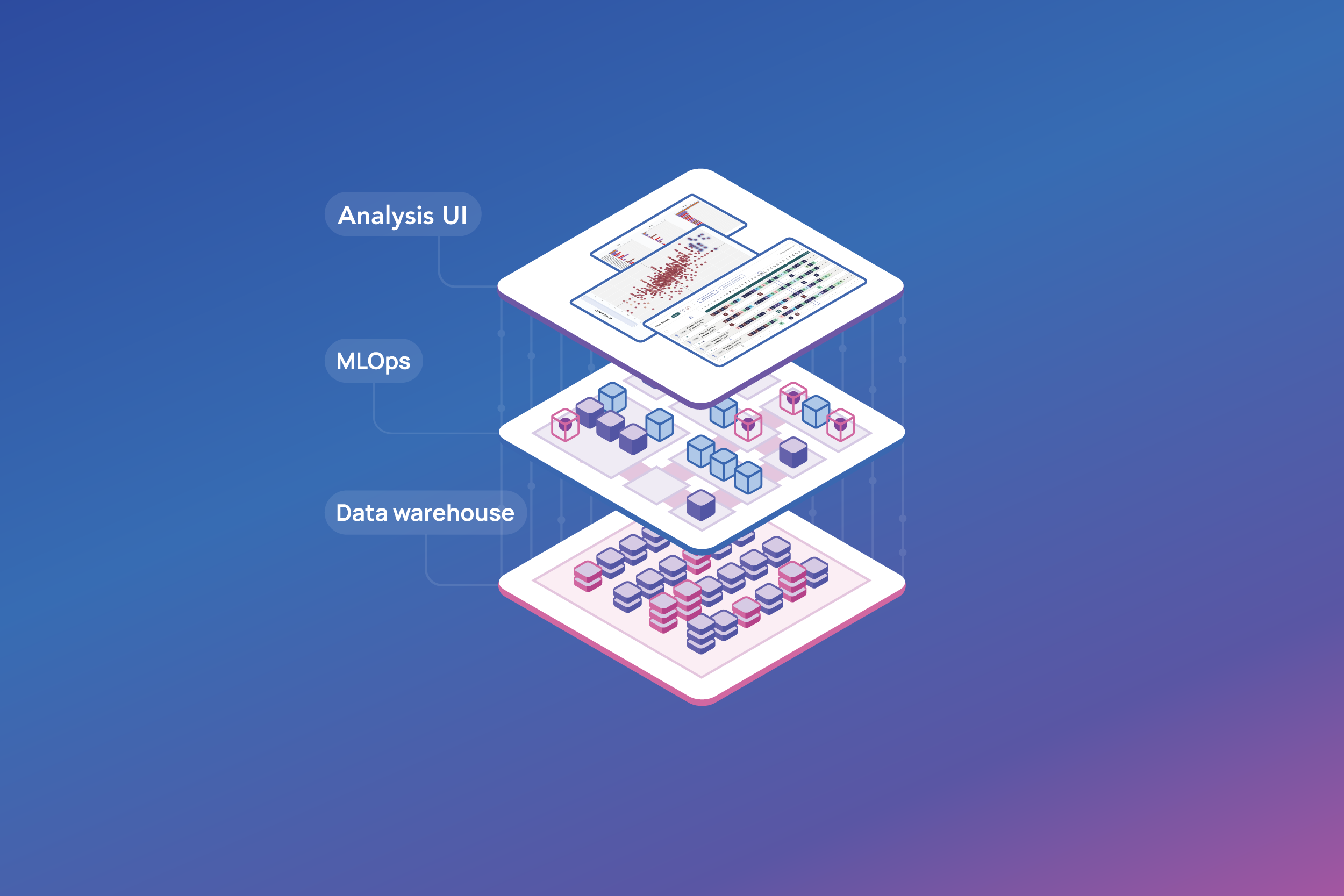
Use case
Quickly register and deploy new models for candidate optimization
In biologics discovery, new AI models are constantly being developed to enhance lead optimization, antibody engineering, and candidate selection.
In this example, we are looking at a candidate optimization model described in this study, that utilizes machine learning algorithms to predict and generate antibody sequences with high binding affinity and diversity.
By learning from existing antibody-antigen interaction data, the model can suggest novel antibody candidates that are more likely to exhibit strong binding properties, accelerating the development of effective therapeutics. While this model is publicly available on GitHub, to be effectively used in biologics discovery, it needs to be trained with target-specific data, validated, and deployed in a structured way so that lab scientists can easily access and apply its predictions.
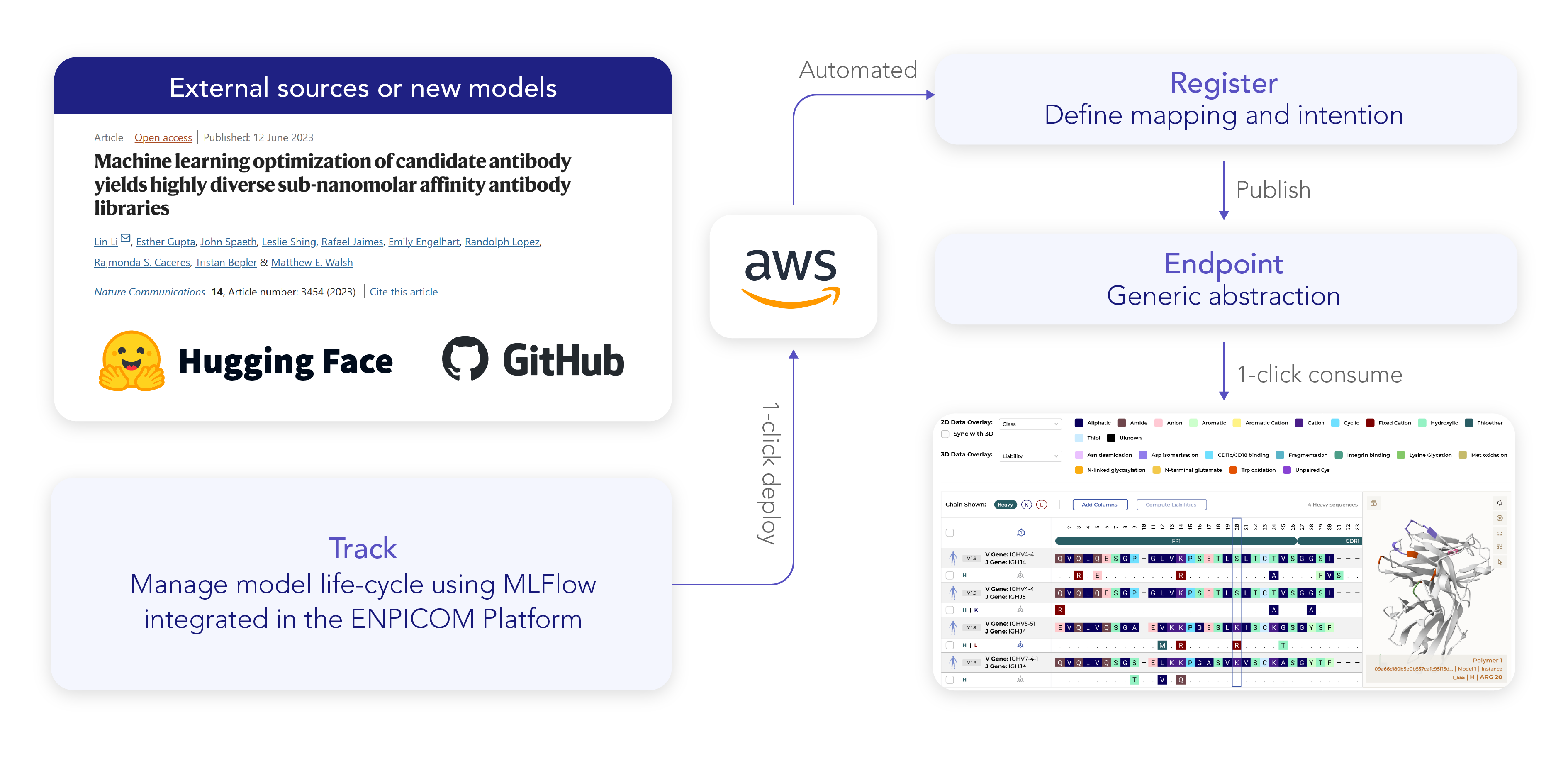
From research paper to implementation in a day
Retrieve the model from external sources
If the model is publicly available on Hugging Face or GitHub, ML scientists download it to their local or cloud environment.
Upload and train the model in MLFlow
- ML scientists retrieve relevant dataset(s) from the database and train the model for specific targets.
- The trained model is then uploaded into the integrated MLFlow environment within the ENPICOM Platform.
- Multiple datasets are tested to compare model quality and evaluate different training strategies.
Track model performance and select the best version
- ML scientists track each trained model’s performance metrics, input data, and results within MLFlow.
- By comparing different versions, they can identify the best-performing model for deployment.
Deploy to CloudOps for automated execution
- Once a final model is selected, it is deployed to CloudOps, ensuring it can be automatically executed without manual intervention.
- CloudOps manages scalability, resource allocation, and uptime, ensuring the model is always available when needed.
Register and define model intent
- The automated model registry logs which version is being deployed, ensuring full traceability.
- The model’s intent is determined, including what type of input data it requires and where it can be accessed in the interface for lab scientists.
- Inputs are mapped for automated data extraction, ensuring lab scientists can use the model without technical setup.
Model becomes available in the right interface
- The model is now accessible to lab scientists in the appropriate section of the ENPICOM Platform.
- In this case, the model becomes available in the Basket environment, where scientists can generate optimized variants of initially selected candidates.
Trusted by industry leaders












Power your ML journey with seamless collaboration between data and lab scientists
Contact us for a personalized demo or consultation.