Every discipline of biomedical research is generating data at an exponential, breakneck pace. Increased experiment throughput, like next-generation sequencing, is creating a wealth of information for scientists. Importantly, this data is the key to discovery—but only if we can properly analyze it.
Before research was inundated with such vast amounts of varied information, data and its analysis tools were tightly linked. Sequencers and mass spectrometers, for example, were controlled by software platforms that also analyzed sequence and spectrum data. Software engineers could also leverage specific data formats to create programs that could, for example, infer gene expression data from a next-generation RNA sequencing experiment. Now, we need to efficiently access huge amounts of different kinds of data to solve complex problems.
Today, data is the key to improving efficiency within companies and academia, saving time and decreasing costs. Automating routine tasks such as data entry, inventory, and supply chain tracking can not only reduce errors but also save time and personnel costs. In the pharmaceutical industry, in particular, software systems that automate quality control and batch tracking can improve drug manufacturing consistency while lowering costs. In order to automate these processes, however, a solution is required that can bridge the divide between relevant data sources and data analysis applications while keeping data secure.
AI has additionally changed how biomedical researchers approach scientific problems. Rather than relying on one large dataset, like the human genome or human proteome, to determine the cause or potential treatments for a disease, deep-learning models can analyze vast, complex, and unrelated datasets, including sequencing data, research papers, chemical library screens, and chemical binding and synthesis data to determine, for example, the most promising leads for drug development.
One of the major hurdles to automating data entry or quality control processes and leveraging deep-learning models for data analysis is the instrument connectivity challenge: connecting siloed data sources with the analytic software tools required to streamline operations or answer complex biological questions. Fortunately, application programming interfaces, or APIs, exist to bridge the gap between siloed data and these critical analysis tools.
What is an API?
APIs are created for one purpose: to allow two or more computer programs to communicate with one another. A good analogy for an API is the wait staff at a restaurant. The customer and chef at any restaurant are separated and cannot speak to one another, much like siloed research data and software analysis tools. But APIs, much like the waiter at a restaurant, can communicate a customer’s order to the chef as a messenger so the chef can make the correct food. Likewise, APIs allow software tools to comb through and analyze data it couldn’t access before (Figure 1).
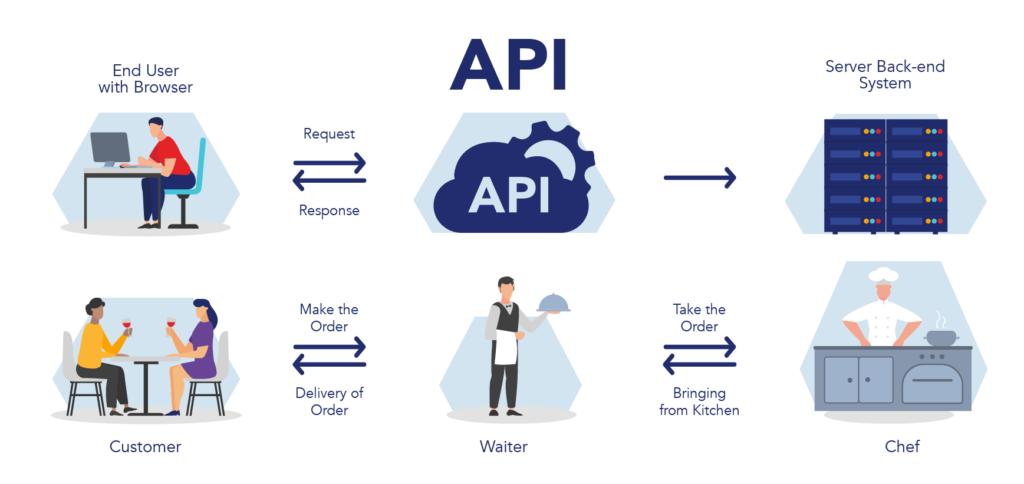
Figure 1. APIs allow communication between two or more software systems, much like a waiter communicating between the customer and the chef at a restaurant.1
Importantly, APIs overcome the major hurdles of big data in biomedical research, commonly referred to as the 4 Vs: volume, velocity, variety, and veracity.2 More specifically, scientific research is currently creating an enormous volume of research data at such a velocity and from a variety of different sources and at varying quality that it is becoming increasingly difficult for researchers to normalize and organize data so that it can be effectively assessed and analyzed to answer real scientific questions or streamline operations.
APIs overcome these challenges to make data accessible and analyze it with the most innovative models and algorithms available. Today, researchers are more aware of current data access issues and format and store their newly generated data according to FAIR data guidelines to ensure the data is findable, accessible, interoperable (works with different systems), and usable for other researchers (Figure 2). APIs, on the other hand, essentially make siloed legacy or proprietary datasets FAIR retroactively, allowing software analysis tools to communicate with large and differing data sources.
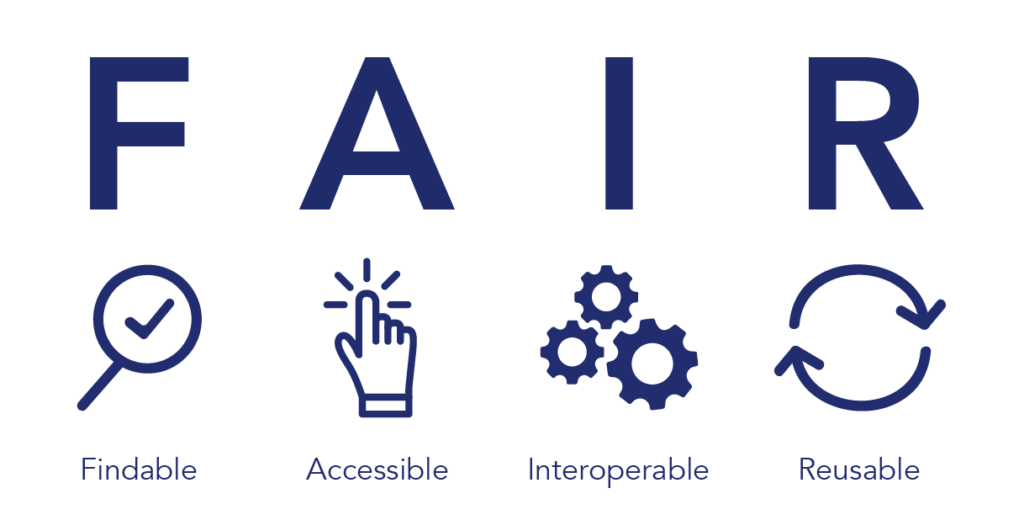
Figure 2. FAIR data standards ensure that data is findable, accessible, interoperable, and reusable to democratize data access.3
APIs in drug discovery
A great deal of scientific discovery is based on detecting natural patterns and exceptions to those patterns. In order to detect patterns, however, researchers require data.
Importantly, the scientific method hasn’t changed. What has changed is the amount of data we are capable of producing and the way in which we analyze it. Data storage and computation hurdles are overcome every day with new and more powerful cloud computing, data storage, and AI solutions. Despite this, even the most sophisticated deep-learning model requires data to train to detect patterns and exceptions to that pattern. The more quality data models receive during training, the more accurately they can describe or predict answers to scientific questions.
One of the most obvious places AI analysis can be applied in biomedical research is in the early stages of drug discovery when several compounds are selected as candidates for disease therapy. Pharmaceutical companies have struggled for decades with identifying compounds or biologicals that have the greatest likelihood for safety and therapeutic success in humans. In fact, upwards of 90% of all drug candidates will fail in human clinical trials,4 representing a major pain point and source of waste in the field.
The application of AI in drug discovery promises to eliminate a good portion of drug candidate failures in the pharmaceutical industry. While savings have yet to be realized, some experts estimate that AI and machine learning (ML) could reduce the time and cost required to develop a drug by 25 to 50 percent.5 Some of the drug discovery challenges AI and machine learning data analysis will address include:
- Target identification — ML models can comb through vast amounts of data to identify new therapeutic targets humans may have missed
- Molecular simulations — Improvements in predictive algorithms can help researchers determine which compounds will bind with the greatest specificity to a therapeutic target
- Translational predictions — Modeling can improve scientists’ ability to predict both the efficacy and toxicity of a drug in humans
- Drug design — Generative AI can suggest new manufacturable compounds based on data used to train learning models
- Candidate prioritization — Algorithms can guide researchers toward candidate drugs with the best potential efficacy and safety profiles based on vast amounts of training data
- Synthesis pathways — AI can help chemical engineers develop high-yield, low-cost synthesis reactions for candidate compounds based on training data
- Potential patients — Software will be able to identify new and existing patient populations that may be helped by a specific drug
Importantly, however, machine and deep-learning tools that leverage AI to discover candidate compounds or biologics for a potential therapy are worthless without quality training data and access to the experimental, clinical, genome, proteome, or other data required to test a scientific hypothesis. While current and future data generation is moving toward FAIR data standardization practices to democratize data access, legacy or proprietary datasets still require an API to communicate between the data sources and the analysis software tool.
Benefits of APIs in drug discovery
ENPICOM’s comprehensive data management and analysis solution, IGX Platform, places great emphasis on API development. This allows drug discovery programs to integrate and manage data sources like electronic laboratory notebooks (ELNs) and laboratory information management systems (LIMSs) to include clone, assay, reliability prediction, sequencing, and other data in a single platform that serves as a launching point for communication with other applications. The platform also mediates the automation of operations, such as data entry and quality control processes, to increase efficiency and decrease errors and costs (Figure 3). The benefits of IGX Platform’s API integration include:
- Synchronization — Keep all laboratory meta-information on a single platform to automatically synchronize experimental data. Labs can also connect to external data storage, such as Amazon S3, to automatically import or export data.
- Automation — Streamline data analysis by creating automated pipelines with an intuitive interface while maintaining the flexibility to explore results and conduct additional case-specific analyses
- Leverage custom code — Run proprietary algorithms to predict candidate compound properties or sequence clustering, make results available to scientists during candidate selection, and craft custom visualizations or analyses with lab data
- Data organization and machine learning — Leverage the scalable architecture and accessible interface of the IGX Platform to easily retrieve datasets to train your in-house machine learning models
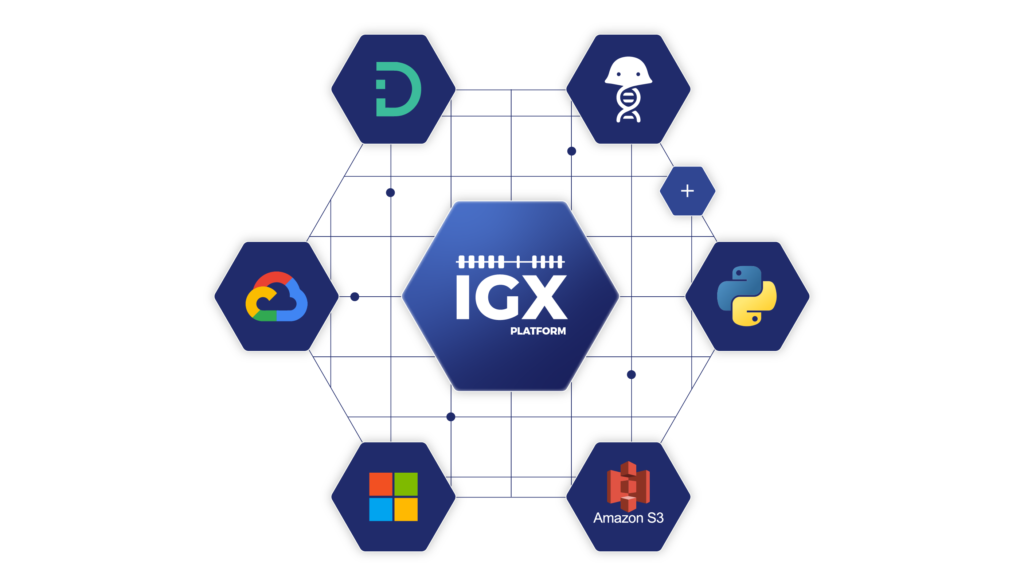
Figure 3. The comprehensive API enables seamless integration with systems like LIMS and ELN, used in therapeutics discovery and development. Using the API SDK, scientists can run custom algorithms outside of the IGX Platform, while also enabling rapid querying of large annotated datasets stored in the platform.
Harnessing the power of big data requires an effective and comprehensive API solution. Visit our technology page to learn how the IGX Platform can facilitate the next great discovery in your research program, or contact us with any questions you have regarding the role of APIs in biomedical research.
References
- What is an API (Application Programming Interface)? (2024). Geeks for Geeks. https://www.geeksforgeeks.org/what-is-an-api/
- Zhu H. (2020). Big Data and Artificial Intelligence Modeling for Drug Discovery. Annual review of pharmacology and toxicology, 60, 573–589. https://doi.org/10.1146/annurev-pharmtox-010919-023324
- Wilkinson, M. D. et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data, 3, 160018. doi:10.1038/sdata.2016.18
- Mullard A. (2016). Parsing Clinical Success Rates. Nature reviews drug discovery, 15, 447. https://doi.org/10.1038/nrd.2016.136
- Unlocking the Potential of AI in Drug Discovery. (2023). The Wellcome Trust. https://cms.wellcome.org/sites/default/files/2023-06/unlocking-the-potential-of-AI-in-drug-discovery_report.pdf