Unlock the full potential of your repertoire data
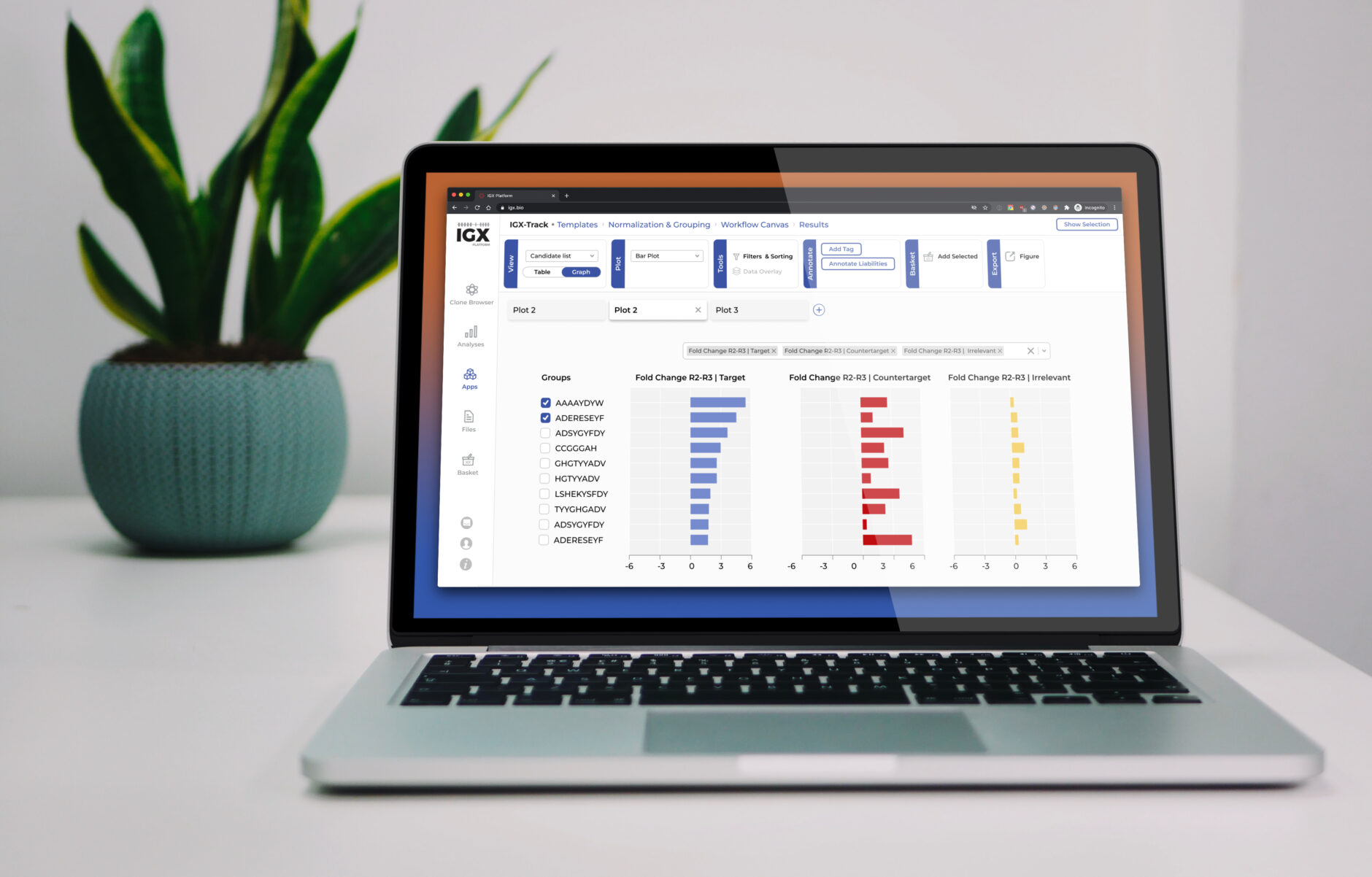
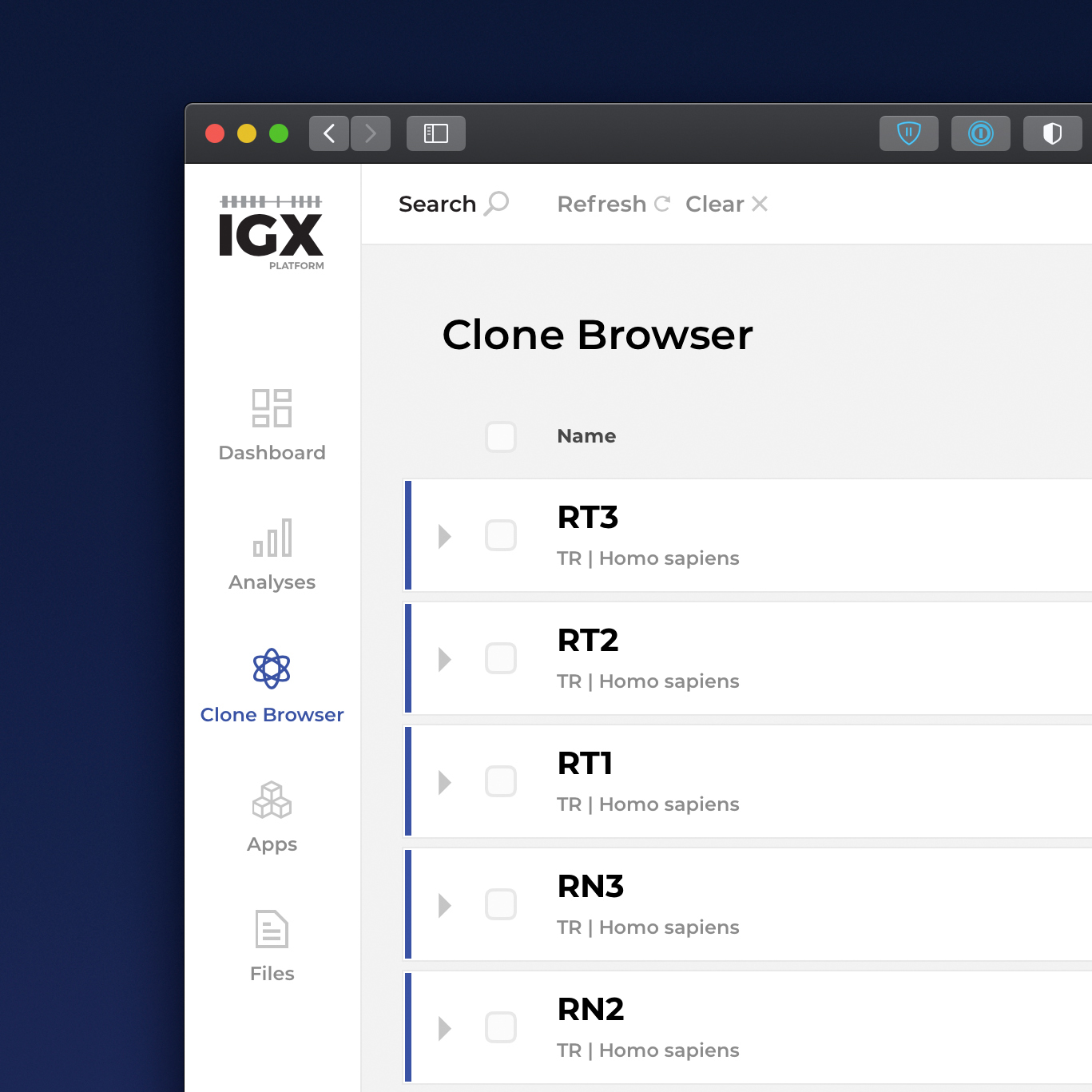
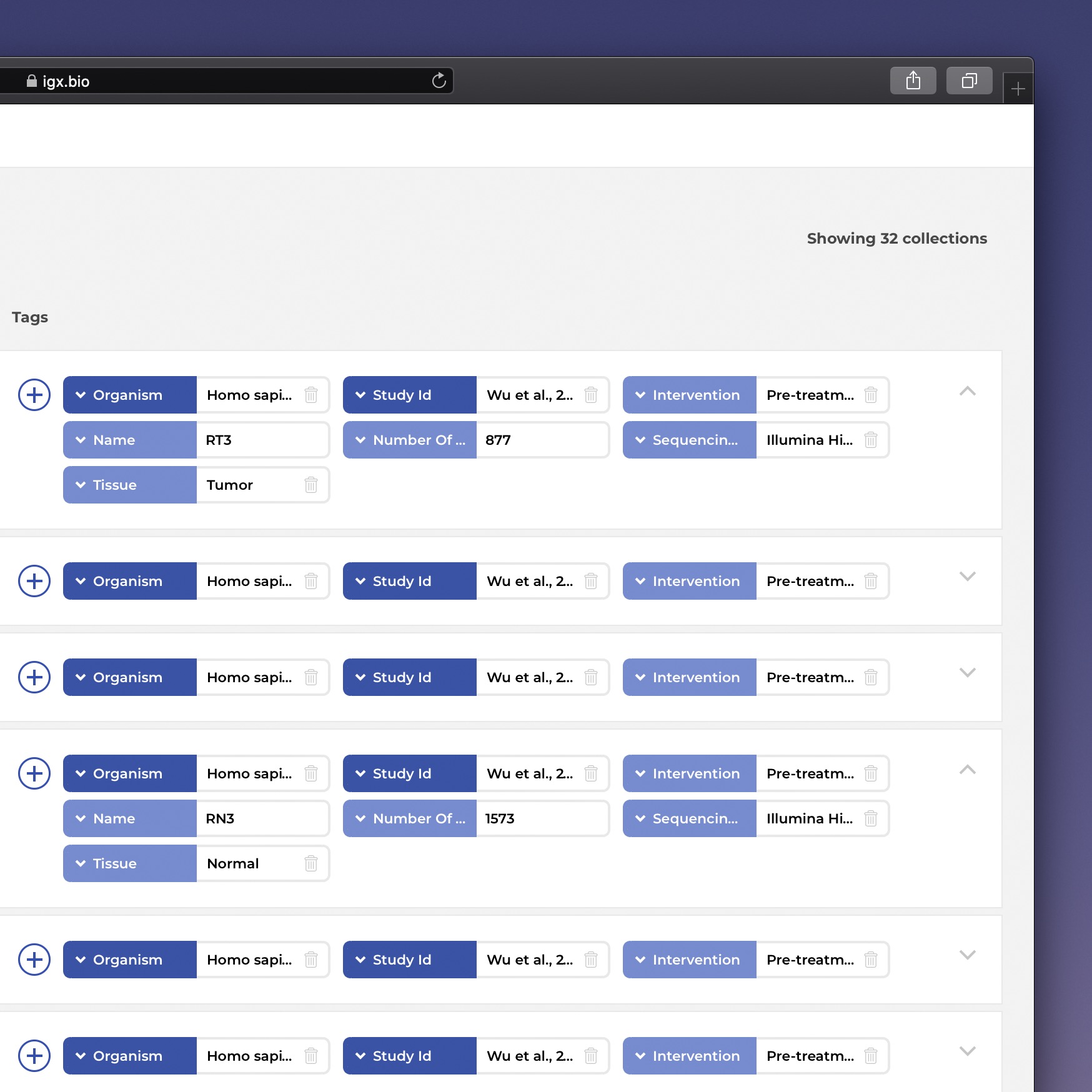
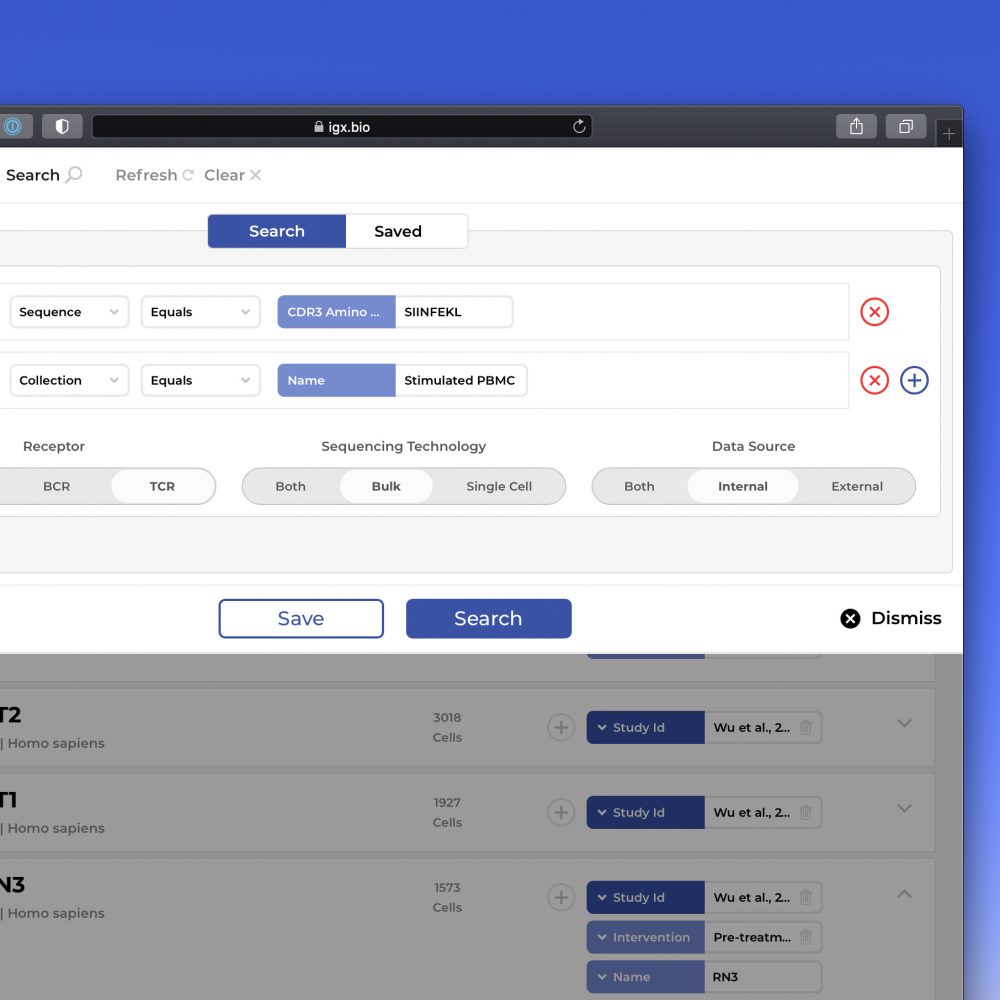
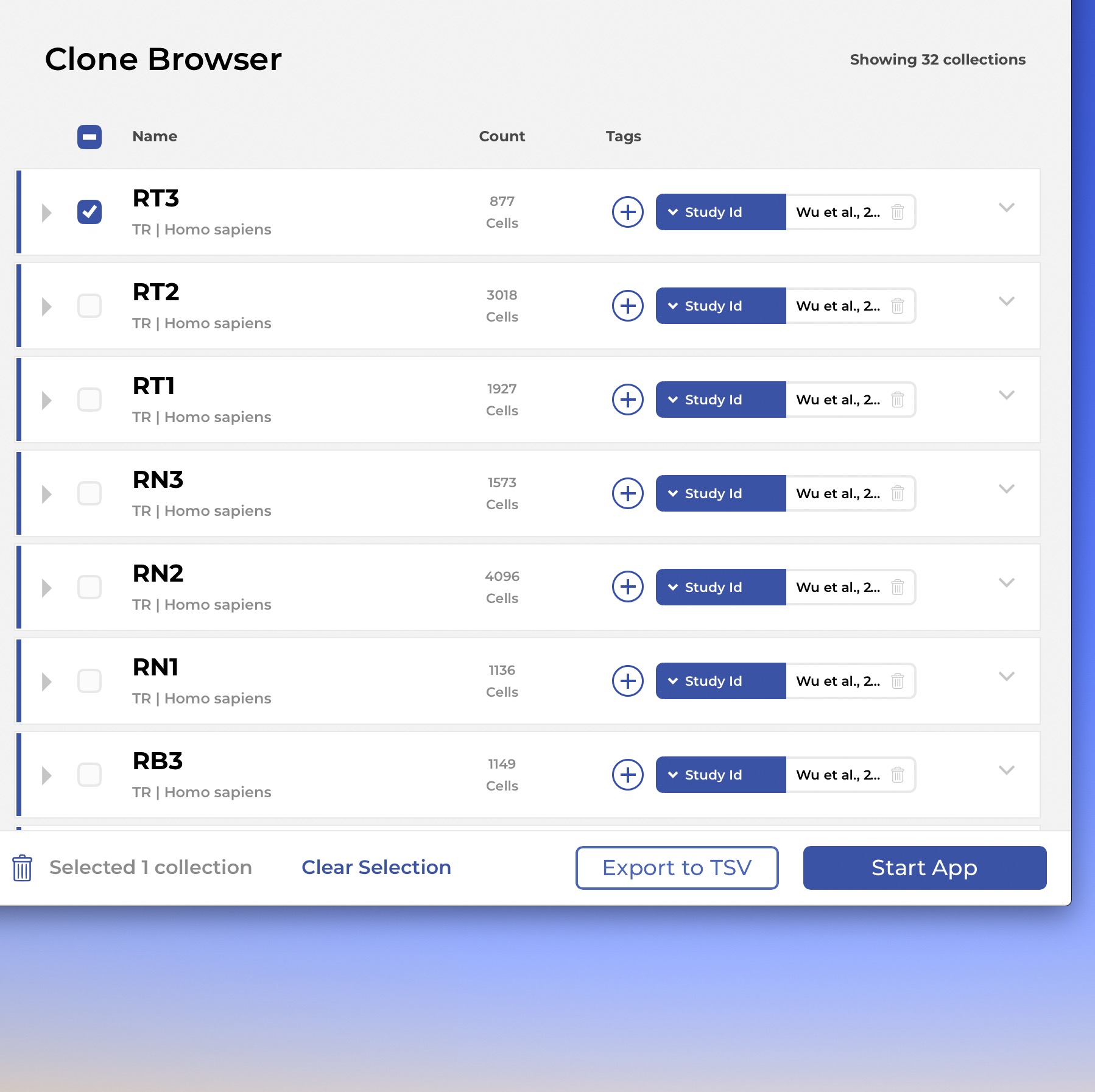
The IGX Platform is an end-to-end cloud solution that removes the complexity of managing and analyzing immune repertoire sequencing data. Technology-agnostic and code-free, it enables every scientist to focus on the research and maximizing valuable insights.
- Work with T and B cells, bulk NGS, Single-cell, and Sanger
- Integrate data from any source
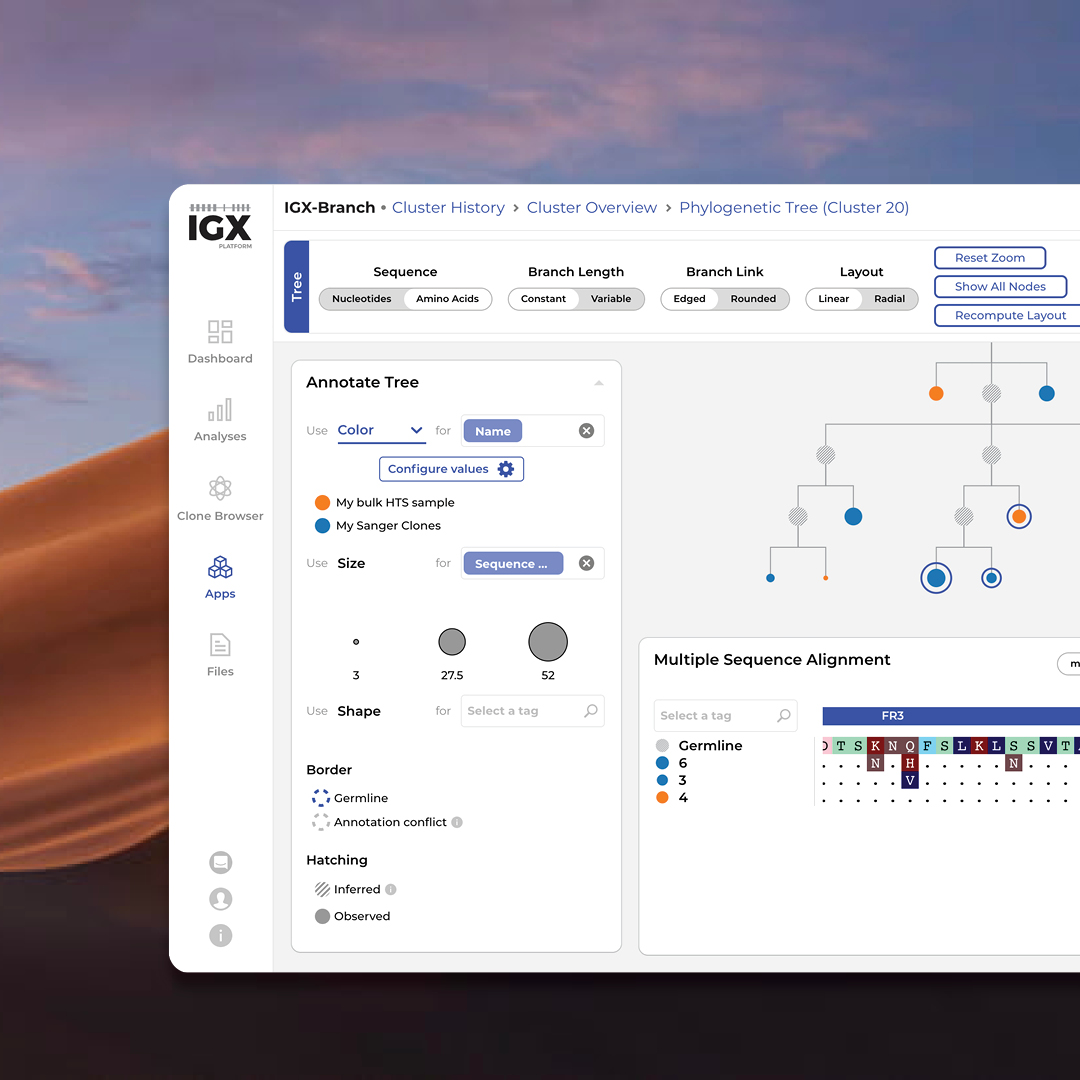
- Intuitive user interface and visualizations
- Always on, ready to scale